Identifying Candidate Targets of Immune Responses in Zika Virus Based on Homology to Epitopes in Other Flavivirus Species
- PMID: 28018746
- PMCID: PMC5145810
- DOI: 10.1371/currents.outbreaks.9aa2e1fb61b0f632f58a098773008c4b
Identifying Candidate Targets of Immune Responses in Zika Virus Based on Homology to Epitopes in Other Flavivirus Species
Abstract
Introduction: The current outbreak of Zika virus has resulted in a massive effort to accelerate the development of ZIKV-specific diagnostics and vaccines. These efforts would benefit greatly from the definition of the specific epitope targets of immune responses in ZIKV, but given the relatively recent emergence of ZIKV as a pandemic threat, few such data are available.
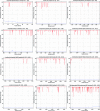
Methods: We used a large body of epitope data for other Flaviviruses that was available from the IEDB for a comparative analysis against the ZIKV proteome in order to project targets of immune responses in ZIKV.
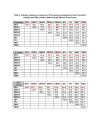
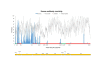
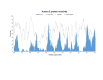
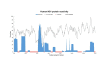
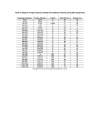
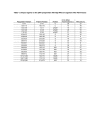
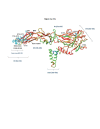
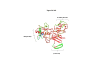
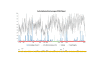
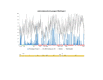
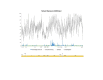
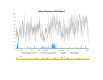
Results: We found a significant level of overlap between known antigenic sites from other Flavivirus proteins with residues on the ZIKV polyprotein. The E and NS1 proteins shared functional antibody epitope sites, whereas regions of T cell reactivity were conserved within NS3 and NS5 for ZIKV. Discussion: Our epitope based analysis provides guidance for which regions of the ZIKV polyprotein are most likely unique targets of ZIKV-specific antibodies, and which targets in ZIKV are most likely to be cross-reactive with other Flavivirus species. These data may therefore provide insights for the development of antibody- and T cell-based ZIKV-specific diagnostics, therapeutics and prophylaxis.
Figures


References
-
- Fauci AS, Morens DM: Zika Virus in the Americas - Yet Another Arbovirus Threat. N Engl J Med 2016, 374:601-604. - PubMed
-
- Driggers RW, Ho CY, Korhonen EM, Kuivanen S, Jääskeläinen AJ, Smura T, Rosenberg A, Hill DA, DeBiasi RL, Vezina G, Timofeev J, Rodriguez FJ, Levanov L, Razak J, Iyengar P, Hennenfent A, Kennedy R, Lanciotti R, du Plessis A, Vapalahti O. Zika Virus Infection with Prolonged Maternal Viremia and Fetal Brain Abnormalities. N Engl J Med. 2016;374(22):2142-2151. - PubMed
-
- Rubin EJ, Greene MF, Baden LR: Zika Virus and Microcephaly. N Engl J Med 2016;374(10):984-985. - PubMed
-
- WHO situation report
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

