Ubiquitination of hnRNPA1 by TRAF6 links chronic innate immune signaling with myelodysplasia
- PMID: 28024152
- PMCID: PMC5423405
- DOI: 10.1038/ni.3654
Ubiquitination of hnRNPA1 by TRAF6 links chronic innate immune signaling with myelodysplasia
Erratum in
-
Corrigendum: Ubiquitination of hnRNPA1 by TRAF6 links chronic innate immune signaling with myelodysplasia.Nat Immunol. 2017 Mar 22;18(4):474. doi: 10.1038/ni0417-474a. Nat Immunol. 2017. PMID: 28323261 Free PMC article. No abstract available.
Abstract
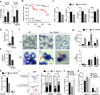
Toll-like receptor (TLR) activation contributes to premalignant hematologic conditions, such as myelodysplastic syndromes (MDS). TRAF6, a TLR effector with ubiquitin (Ub) ligase activity, is overexpressed in MDS hematopoietic stem/progenitor cells (HSPCs). We found that TRAF6 overexpression in mouse HSPC results in impaired hematopoiesis and bone marrow failure. Using a global Ub screen, we identified hnRNPA1, an RNA-binding protein and auxiliary splicing factor, as a substrate of TRAF6. TRAF6 ubiquitination of hnRNPA1 regulated alternative splicing of Arhgap1, which resulted in activation of the GTP-binding Rho family protein Cdc42 and accounted for hematopoietic defects in TRAF6-expressing HSPCs. These results implicate Ub signaling in coordinating RNA processing by TLR pathways during an immune response and in premalignant hematologic diseases, such as MDS.
Figures


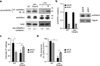




Comment in
-
RNA-binding proteins mind the GAPs.Nat Immunol. 2017 Jan 19;18(2):146-148. doi: 10.1038/ni.3662. Nat Immunol. 2017. PMID: 28102216 No abstract available.
-
Chronic innate immune signaling results in ubiquitination of splicing machinery.Cell Cycle. 2018;17(4):407-409. doi: 10.1080/15384101.2018.1429082. Epub 2018 Apr 3. Cell Cycle. 2018. PMID: 29336715 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

