Alternative splicing in tomato pollen in response to heat stress
- PMID: 28025318
- PMCID: PMC5397606
- DOI: 10.1093/dnares/dsw051
Alternative splicing in tomato pollen in response to heat stress
Abstract
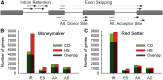
Alternative splicing (AS) is a key control mechanism influencing signal response cascades in different developmental stages and under stress conditions. In this study, we examined heat stress (HS)-induced AS in the heat sensitive pollen tissue of two tomato cultivars. To obtain the entire spectrum of HS-related AS, samples taken directly after HS and after recovery were combined and analysed by RNA-seq. For nearly 9,200 genes per cultivar, we observed at least one AS event under HS. In comparison to control, for one cultivar we observed 76% more genes with intron retention (IR) or exon skipping (ES) under HS. Furthermore, 2,343 genes had at least one transcript with IR or ES accumulated under HS in both cultivars. These genes are involved in biological processes like protein folding, gene expression and heat response. Transcriptome assembly of these genes revealed that most of the alternative spliced transcripts possess truncated coding sequences resulting in partial or total loss of functional domains. Moreover, 141 HS specific and 22 HS repressed transcripts were identified. Further on, we propose AS as layer of stress response regulating constitutively expressed genes under HS by isoform abundance.
Keywords: RNA-seq; alternative splicing; assembly; heat stress; tomato pollen; transcriptome.
© The Author 2016. Published by Oxford University Press on behalf of Kazusa DNA Research Institute.
Figures





References
-
- Fragkostefanakis S., Simm S., Paul P., Bublak D., Scharf K. D., Schleiff E.. 2015, Chaperone network composition in Solanum lycopersicum explored by transcriptome profiling and microarray meta-analysis. Plant Cell Environ., 38, 693–709. - PubMed
-
- Jung K.H., Ko H.J., Nguyen M.X., Kim S.R., Ronald P., An G.. 2012, Genome-wide identification and analysis of early heat stress responsive genes in rice. J. Plant Biol., 55, 10.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

