GeneBase 1.1: a tool to summarize data from NCBI gene datasets and its application to an update of human gene statistics
- PMID: 28025344
- PMCID: PMC5199132
- DOI: 10.1093/database/baw153
GeneBase 1.1: a tool to summarize data from NCBI gene datasets and its application to an update of human gene statistics
Abstract
We release GeneBase 1.1, a local tool with a graphical interface useful for parsing, structuring and indexing data from the National Center for Biotechnology Information (NCBI) Gene data bank. Compared to its predecessor GeneBase (1.0), GeneBase 1.1 now allows dynamic calculation and summarization in terms of median, mean, standard deviation and total for many quantitative parameters associated with genes, gene transcripts and gene features (exons, introns, coding sequences, untranslated regions). GeneBase 1.1 thus offers the opportunity to perform analyses of the main gene structure parameters also following the search for any set of genes with the desired characteristics, allowing unique functionalities not provided by the NCBI Gene itself. In order to show the potential of our tool for local parsing, structuring and dynamic summarizing of publicly available databases for data retrieval, analysis and testing of biological hypotheses, we provide as a sample application a revised set of statistics for human nuclear genes, gene transcripts and gene features. In contrast with previous estimations strongly underestimating the length of human genes, a 'mean' human protein-coding gene is 67 kbp long, has eleven 309 bp long exons and ten 6355 bp long introns. Median, mean and extreme values are provided for many other features offering an updated reference source for human genome studies, data useful to set parameters for bioinformatic tools and interesting clues to the biomedical meaning of the gene features themselves.Database URL: http://apollo11.isto.unibo.it/software/.
© The Author(s) 2016. Published by Oxford University Press.
Figures
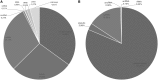
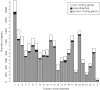
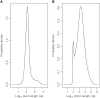
References
-
- Piovesan A., Vitale L., Pelleri M.C. et al. (2013) Universal tight correlation of codon bias and pool of RNA codons (codonome): the genome is optimized to allow any distribution of gene expression values in the transcriptome from bacteria to humans. Genomics, 101, 282–289. - PubMed
-
- Vitale L., Lenzi L., Huntsman S.A. et al. (2006) Differential expression of alternatively spliced mRNA forms of the insulin-like growth factor 1 receptor in human neuroendocrine tumors. Oncol. Rep., 15, 1249–1256. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

