Collective Resistance in Microbial Communities by Intracellular Antibiotic Deactivation
- PMID: 28027306
- PMCID: PMC5189934
- DOI: 10.1371/journal.pbio.2000631
Collective Resistance in Microbial Communities by Intracellular Antibiotic Deactivation
Abstract
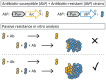
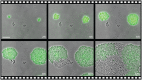
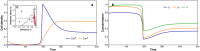
The structure and composition of bacterial communities can compromise antibiotic efficacy. For example, the secretion of β-lactamase by individual bacteria provides passive resistance for all residents within a polymicrobial environment. Here, we uncover that collective resistance can also develop via intracellular antibiotic deactivation. Real-time luminescence measurements and single-cell analysis demonstrate that the opportunistic human pathogen Streptococcus pneumoniae grows in medium supplemented with chloramphenicol (Cm) when resistant bacteria expressing Cm acetyltransferase (CAT) are present. We show that CAT processes Cm intracellularly but not extracellularly. In a mouse pneumonia model, more susceptible pneumococci survive Cm treatment when coinfected with a CAT-expressing strain. Mathematical modeling predicts that stable coexistence is only possible when antibiotic resistance comes at a fitness cost. Strikingly, CAT-expressing pneumococci in mouse lungs were outcompeted by susceptible cells even during Cm treatment. Our results highlight the importance of the microbial context during infectious disease as a potential complicating factor to antibiotic therapy.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- World Health Organization. Antimicrobial resistance: global report on surveillance. 2014 http://www.who.int/drugresistance/documents/surveillancereport/en.
-
- Chi DH, Hendley JO, French P, Arango P, Hayden FG, Winther B. Nasopharyngeal reservoir of bacterial otitis media and sinusitis pathogens in adults during wellness and viral respiratory illness. Am J Rhinol. 2003. August;17(4):209–14. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

