Non-human Primate Schlafen11 Inhibits Production of Both Host and Viral Proteins
- PMID: 28027315
- PMCID: PMC5189954
- DOI: 10.1371/journal.ppat.1006066
Non-human Primate Schlafen11 Inhibits Production of Both Host and Viral Proteins
Abstract
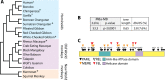
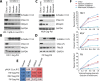
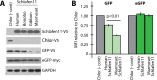
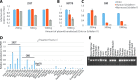
Schlafen11 (encoded by the SLFN11 gene) has been shown to inhibit the accumulation of HIV-1 proteins. We show that the SLFN11 gene is under positive selection in simian primates and is species-specific in its activity against HIV-1. The activity of human Schlafen11 is relatively weak compared to that of some other primate versions of this protein, with the versions encoded by chimpanzee, orangutan, gibbon, and marmoset being particularly potent inhibitors of HIV-1 protein production. Interestingly, we find that Schlafen11 is functional in the absence of infection and reduces protein production from certain non-viral (GFP) and even host (Vinculin and GAPDH) transcripts. This suggests that Schlafen11 may just generally block protein production from non-codon optimized transcripts. Because Schlafen11 is an interferon-stimulated gene with a broad ability to inhibit protein production from many host and viral transcripts, its role may be to create a general antiviral state in the cell. Interestingly, the strong inhibitors such as marmoset Schlafen11 consistently block protein production better than weak primate Schlafen11 proteins, regardless of the virus or host target being analyzed. Further, we show that the residues to which species-specific differences in Schlafen11 potency map are distinct from residues that have been targeted by positive selection. We speculate that the positive selection of SLFN11 could have been driven by a number of different factors, including interaction with one or more viral antagonists that have yet to be identified.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

