Novel Chemical Scaffolds for Inhibition of Rifamycin-Resistant RNA Polymerase Discovered from High-Throughput Screening
- PMID: 28027449
- PMCID: PMC5323270
- DOI: 10.1177/2472555216679994
Novel Chemical Scaffolds for Inhibition of Rifamycin-Resistant RNA Polymerase Discovered from High-Throughput Screening
Abstract
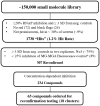
Rifampin has been a cornerstone of tuberculosis (TB) treatment since its introduction. The rise of multidrug-resistant and extensively drug-resistant TB makes the development of novel therapeutics effective against these strains an urgent need. Site-specific mutations in the target enzyme of rifampin, RNA polymerase (RNAP) comprises the majority (~97%) of rifamycin-resistant (RifR) strains of Mycobacterium tuberculosis (MTB). To identify novel inhibitors of bacterial RNAP, an in vitro plasmid-based transcription assay that uses malachite green (MG) to detect transcribed RNA containing MG aptamers was developed. This assay was optimized in a 384-well plate format and used to screen 150,000 compounds against an Escherichia coli homolog of the most clinically relevant RifR RNAP (βS531L) containing a mutation (β'V408G) that compensates for the fitness defect of this RifR mutant. Following confirmation and concentration-response studies, 10 compounds were identified with similar in vitro inhibition values across a panel of wild-type and RifR E. coli and MTB RNAPs. Four compounds identified from the screen are active against MTB in culture at concentrations below 200 µM. Initial follow-up has resulted in the elimination of one scaffold due to potential pan-assay interference.
Keywords: RNA polymerase; antibiotic resistance; high-throughput screen; rifampin; tuberculosis.
Figures

Similar articles
-
Rifamycin inhibition of WT and Rif-resistant Mycobacterium tuberculosis and Escherichia coli RNA polymerases in vitro.Tuberculosis (Edinb). 2011 Sep;91(5):361-9. doi: 10.1016/j.tube.2011.05.002. Epub 2011 Jun 24. Tuberculosis (Edinb). 2011. PMID: 21704562
-
Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism.Nat Commun. 2018 Oct 8;9(1):4147. doi: 10.1038/s41467-018-06587-2. Nat Commun. 2018. PMID: 30297823 Free PMC article.
-
Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition.Mol Cell. 2017 Apr 20;66(2):169-179.e8. doi: 10.1016/j.molcel.2017.03.001. Epub 2017 Apr 6. Mol Cell. 2017. PMID: 28392175 Free PMC article.
-
Resistance to rifampicin: a review.J Antibiot (Tokyo). 2014 Sep;67(9):625-30. doi: 10.1038/ja.2014.107. Epub 2014 Aug 13. J Antibiot (Tokyo). 2014. PMID: 25118103 Review.
-
Bacterial RNA polymerase: a promising target for the discovery of new antimicrobial agents.Curr Opin Investig Drugs. 2007 Aug;8(8):600-7. Curr Opin Investig Drugs. 2007. PMID: 17668362 Review.
Cited by
-
Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis.Mol Microbiol. 2017 Mar;103(6):1034-1045. doi: 10.1111/mmi.13606. Epub 2017 Jan 10. Mol Microbiol. 2017. PMID: 28009073 Free PMC article.
-
The War against Tuberculosis: A Review of Natural Compounds and Their Derivatives.Molecules. 2020 Jun 30;25(13):3011. doi: 10.3390/molecules25133011. Molecules. 2020. PMID: 32630150 Free PMC article. Review.
-
Molecular basis for RNA polymerase-dependent transcription complex recycling by the helicase-like motor protein HelD.Nat Commun. 2020 Dec 18;11(1):6420. doi: 10.1038/s41467-020-20157-5. Nat Commun. 2020. PMID: 33339820 Free PMC article.
-
Discovery of pyrazolopyrrolidinones as potent, broad-spectrum inhibitors of Leishmania infection.Front Trop Dis. 2023;3:1011124. doi: 10.3389/fitd.2022.1011124. Epub 2023 Jan 23. Front Trop Dis. 2023. PMID: 36818551 Free PMC article.
-
Imaging and Tracking RNA in Live Mammalian Cells via Fluorogenic Photoaffinity Labeling.ACS Chem Biol. 2025 Mar 21;20(3):707-720. doi: 10.1021/acschembio.4c00848. Epub 2025 Feb 15. ACS Chem Biol. 2025. PMID: 39953970
References
-
- UNAIDS. Fact Sheet 2016. UNAIDS; 2016. http://www.unaids.org/
-
- World Health Organization. Global Tuberculosis Report 2016. World Health Organization; Geneva, Switzerland: 2016.
-
- Mukhtar TA, Wright GD. Streptogramins, oxazolidinones, and other inhibitors of bacterial protein synthesis. Chem Rev. 2005;105(2):529–542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

