Characterization of founder viruses in very early SIV rectal transmission
- PMID: 28027479
- PMCID: PMC5276734
- DOI: 10.1016/j.virol.2016.12.018
Characterization of founder viruses in very early SIV rectal transmission
Abstract
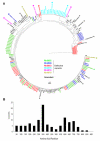
A better understanding of HIV-1 transmission is critical for developing preventative strategies. To that end, we analyzed 524 full-length env sequences of SIVmac251 at 6 and 10 days post intrarectal infection of rhesus macaques. There was no tissue compartmentalization of founder viruses across plasma, rectal and distal lymphatic tissues for most animals; however one animal has evidence of virus tissue compartmentalization. Despite identical viral inoculums, founder viruses were animal-specific, primarily derived from rare variants in the inoculum, and have a founder virus signature that can distinguish dominant founder variants from minor founder or untransmitted variants in the inoculum. Importantly, the sequences of post-transmission defective viruses were phylogenetically associated with competent viral variants in the inoculum and were mainly converted from competent viral variants by frameshift rather than APOBEC mediated mutations, suggesting the converting the transmitted viruses into defective viruses through frameshift mutation is an important component of rectal transmission bottleneck.
Significance: Anorectal receptive intercourse is a common route of HIV-1 transmission and a better understanding of the transmission mechanisms is critical for developing HIV-1 preventative strategies. Here, we report that there is no tissue compartmentalization of founder viruses during very early rectal transmission of SIV in the majority of rhesus macaques and founder viruses are preferentially derived from rare variant in the inoculum. We also found that founder viruses are animal-specific despite identical viral inoculums. After viruses cross the mucosal barriers, the host further reduces viral diversity by converting some of the transmitted functional viruses into defective viruses through frameshift rather than APOBEC derived mutations. To our knowledge, this is the first study of founder viruses at multiple tissue sites during very early rectal transmission.
Keywords: Defective variants; Founder virus signature; Genetic bottleneck; Rectal transmission; Tissue compartmentalization; Very early virological events.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

