Analyses of differentially expressed genes after exposure to acute stress, acute ethanol, or a combination of both in mice
- PMID: 28027852
- PMCID: PMC5270764
- DOI: 10.1016/j.alcohol.2016.08.008
Analyses of differentially expressed genes after exposure to acute stress, acute ethanol, or a combination of both in mice
Abstract
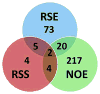
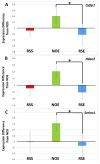
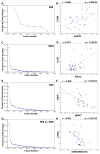
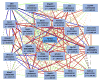
Alcohol abuse is a complex disorder, which is confounded by other factors, including stress. In the present study, we examined gene expression in the hippocampus of BXD recombinant inbred mice after exposure to ethanol (NOE), stress (RSS), and the combination of both (RSE). Mice were given an intraperitoneal (i.p.) injection of 1.8 g/kg ethanol or saline, and subsets of both groups were exposed to acute restraint stress for 15 min or controls. Gene expression in the hippocampus was examined using microarray analysis. Genes that were significantly (p < 0.05, q < 0.1) differentially expressed were further evaluated. Bioinformatic analyses were predominantly performed using tools available at GeneNetwork.org, and included gene ontology, presence of cis-regulation or polymorphisms, phenotype correlations, and principal component analyses. Comparisons of differential gene expression between groups showed little overlap. Gene Ontology demonstrated distinct biological processes in each group with the combined exposure (RSE) being unique from either the ethanol (NOE) or stress (RSS) group, suggesting that the interaction between these variables is mediated through diverse molecular pathways. This supports the hypothesis that exposure to stress alters ethanol-induced gene expression changes and that exposure to alcohol alters stress-induced gene expression changes. Behavior was profiled in all groups following treatment, and many of the differentially expressed genes are correlated with behavioral variation within experimental groups. Interestingly, in each group several genes were correlated with the same phenotype, suggesting that these genes are the potential origins of significant genetic networks. The distinct sets of differentially expressed genes within each group provide the basis for identifying molecular networks that may aid in understanding the complex interactions between stress and ethanol, and potentially provide relevant therapeutic targets. Using Ptp4a1, a candidate gene underlying the quantitative trait locus for several of these phenotypes, and network analyses, we show that a large group of differentially expressed genes in the NOE group are highly interrelated, some of which have previously been linked to alcohol addiction or alcohol-related phenotypes.
Keywords: Bioinformatic analysis; Ethanol; Gene expression; Hippocampus; Stress.
Published by Elsevier Inc.
Figures

References
-
- Alfonso J, Frasch AC, Flugge G. Chronic stress, depression and anti-depressants: Effects on gene transcription in the hippocampus. Reviews in the Neurosciences. 2005;16:43–56. - PubMed
-
- Becker HC, Lopez MF. Increased ethanol drinking after repeated chronic ethanol exposure and withdrawal experience in C57BL/6 mice. Alcoholism: Clinical and Experimental Research. 2004;28:1829–1838. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

