Genetically engineered mouse models in oncology research and cancer medicine
- PMID: 28028012
- PMCID: PMC5286388
- DOI: 10.15252/emmm.201606857
Genetically engineered mouse models in oncology research and cancer medicine
Abstract
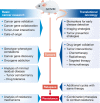
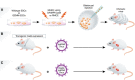
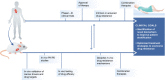
Genetically engineered mouse models (GEMMs) have contributed significantly to the field of cancer research. In contrast to cancer cell inoculation models, GEMMs develop de novo tumors in a natural immune-proficient microenvironment. Tumors arising in advanced GEMMs closely mimic the histopathological and molecular features of their human counterparts, display genetic heterogeneity, and are able to spontaneously progress toward metastatic disease. As such, GEMMs are generally superior to cancer cell inoculation models, which show no or limited heterogeneity and are often metastatic from the start. Given that GEMMs capture both tumor cell-intrinsic and cell-extrinsic factors that drive de novo tumor initiation and progression toward metastatic disease, these models are indispensable for preclinical research. GEMMs have successfully been used to validate candidate cancer genes and drug targets, assess therapy efficacy, dissect the impact of the tumor microenvironment, and evaluate mechanisms of drug resistance. In vivo validation of candidate cancer genes and therapeutic targets is further accelerated by recent advances in genetic engineering that enable fast-track generation and fine-tuning of GEMMs to more closely resemble human patients. In addition, aligning preclinical tumor intervention studies in advanced GEMMs with clinical studies in patients is expected to accelerate the development of novel therapeutic strategies and their translation into the clinic.
Keywords: cancer; genetically engineered mouse models; metastasis; therapy; tumor microenvironment.
© 2016 The Authors. Published under the terms of the CC BY 4.0 license.
Figures


References
-
- Bald T, Quast T, Landsberg J, Rogava M, Glodde N, Lopez‐Ramos D, Kohlmeyer J, Riesenberg S, van den Boorn‐Konijnenberg D, Hömig‐Hölzel C et al (2014) Ultraviolet‐radiation‐induced inflammation promotes angiotropism and metastasis in melanoma. Nature 507: 109–113 - PubMed
-
- Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C, Corà D, Di Nicolantonio F, Buscarino M, Petti C et al (2011) A molecularly annotated platform of patient‐derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab‐resistant colorectal cancer. Cancer Discov 1: 508–523 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

