The Multiple Roles of Exosomes in Metastasis
- PMID: 28031234
- PMCID: PMC5267497
- DOI: 10.21873/cgp.20015
The Multiple Roles of Exosomes in Metastasis
Abstract
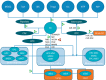
Exosomes are important contributors to cell-cell communication and their role as diagnostic markers for cancer and the pathogenesis for cancer is under intensive investigation. Here, we focus on their role in metastasis-related processes. We discuss their impact regarding promotion of invasion and migration of tumor cells, conditioning of lymph nodes, generation of premetastatic niches and organotropism of metastasis. Furthermore, we highlight interactions of exosomes with bone marrow and stromal components such as fibroblasts, endothelial cells, myeloid- and other immune-related cells in the context of metastases. For all processes as described above, we outline molecular and cellular components for therapeutic intervention with metastatic processes.
Keywords: Exosome interaction with stromal cells; organ tropism of metastasis; pre- and metastatic niche; review.
Copyright© 2017, International Institute of Anticancer Research (Dr. George J. Delinasios), All rights reserved.
Figures
References
-
- Langley RR, Fidler IJ. Tumor cell-organ microenvironment interactions in the pathogenesis of cancer metastases. Endocr Reviews. 2007;28:297–321. - PubMed
-
- Hüsemann Y, Geigl JB, Schubert F, Meyer M, Burghart E, Forni G, Eils R, Fehm T, Rietmüller G, Klein CA. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
