High-throughput identification of C/D box snoRNA targets with CLIP and RiboMeth-seq
- PMID: 28031372
- PMCID: PMC5389715
- DOI: 10.1093/nar/gkw1321
High-throughput identification of C/D box snoRNA targets with CLIP and RiboMeth-seq
Abstract
High-throughput sequencing has greatly facilitated the discovery of long and short non-coding RNAs (ncRNAs), which frequently guide ribonucleoprotein complexes to RNA targets, to modulate their metabolism and expression. However, for many ncRNAs, the targets remain to be discovered. In this study, we developed computational methods to map C/D box snoRNA target sites using data from core small nucleolar ribonucleoprotein crosslinking and immunoprecipitation and from transcriptome-wide mapping of 2΄-O-ribose methylation sites. We thereby assigned the snoRNA guide to a known methylation site in the 18S rRNA, we uncovered a novel partially methylated site in the 28S ribosomal RNA, and we captured a site in the 28S rRNA in interaction with multiple snoRNAs. Although we also captured mRNAs in interaction with snoRNAs, we did not detect 2΄-O-methylation of these targets. Our study provides an integrated approach to the comprehensive characterization of 2΄-O-methylation targets of snoRNAs in species beyond those in which these interactions have been traditionally studied and contributes to the rapidly developing field of 'epitranscriptomics'.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
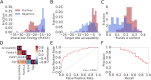
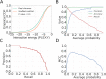


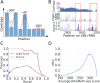
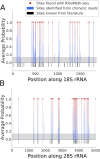
References
-
- Li S., Mason C.E.. The pivotal regulatory landscape of RNA modifications. Annu. Rev. Genomics Hum. Genet. 2014; 15:127–150. - PubMed
-
- Birkedal U., Christensen-Dalsgaard M., Krogh N., Sabarinathan R., Gorodkin J., Nielsen H.. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew. Chem. Int. Ed. Engl. 2014; 10.1002/anie.201408362. - PubMed
-
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. - PubMed
-
- Schaefer M. RNA 5-methylcytosine analysis by bisulfite sequencing. Methods Enzymol. 2015; 560:297–329. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

