PRMT5 C-terminal Phosphorylation Modulates a 14-3-3/PDZ Interaction Switch
- PMID: 28031468
- PMCID: PMC5313098
- DOI: 10.1074/jbc.M116.760330
PRMT5 C-terminal Phosphorylation Modulates a 14-3-3/PDZ Interaction Switch
Abstract
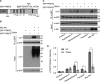
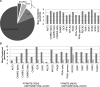
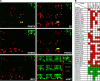
PRMT5 is the primary enzyme responsible for the deposition of the symmetric dimethylarginine in mammalian cells. In an effort to understand how PRMT5 is regulated, we identified a threonine phosphorylation site within a C-terminal tail motif, which is targeted by the Akt/serum- and glucocorticoid-inducible kinases. While investigating the function of this posttranslational modification, we serendipitously discovered that its free C-terminal tail binds PDZ domains (when unphosphorylated) and 14-3-3 proteins (when phosphorylated). In essence, a phosphorylation event within the last few residues of the C-terminal tail generates a posttranslational modification-dependent PDZ/14-3-3 interaction "switch." The C-terminal motif of PRMT5 is required for plasma membrane association, and loss of this switching capacity is not compatible with life. This signaling phenomenon was recently reported for the HPV E6 oncoprotein but has not yet been observed for mammalian proteins. To investigate the prevalence of PDZ/14-3-3 switching in signal transduction, we built a protein domain microarray that harbors PDZ domains and 14-3-3 proteins. We have used this microarray to interrogate the C-terminal tails of a small group of candidate proteins and identified ERBB4, PGHS2, and IRK1 (as well as E6 and PRMT5) as conforming to this signaling mode, suggesting that PDZ/14-3-3 switching may be a broad biological paradigm.
Keywords: 14–3-3 protein; PDZ domain; cell signaling; protein arginine N-methyltransferase 5 (PRMT5); protein methylation; protein phosphorylation.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

