Identification of NAD+ capped mRNAs in Saccharomyces cerevisiae
- PMID: 28031484
- PMCID: PMC5255579
- DOI: 10.1073/pnas.1619369114
Identification of NAD+ capped mRNAs in Saccharomyces cerevisiae
Abstract
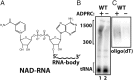
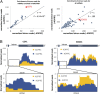
RNAs besides tRNA and rRNA contain chemical modifications, including the recently described 5' nicotinamide-adenine dinucleotide (NAD+) RNA in bacteria. Whether 5' NAD-RNA exists in eukaryotes remains unknown. We demonstrate that 5' NAD-RNA is found on subsets of nuclear and mitochondrial encoded mRNAs in Saccharomyces cerevisiae NAD-mRNA appears to be produced cotranscriptionally because NAD-RNA is also found on pre-mRNAs, and only on mitochondrial transcripts that are not 5' end processed. These results define an additional 5' RNA cap structure in eukaryotes and raise the possibility that this 5' NAD+ cap could modulate RNA stability and translation on specific subclasses of mRNAs.
Keywords: NAD-RNA; RNA modification; mitochondria; transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

