Spatially-dependent Bayesian model selection for disease mapping
- PMID: 28034176
- PMCID: PMC5374035
- DOI: 10.1177/0962280215627298
Spatially-dependent Bayesian model selection for disease mapping
Abstract
In disease mapping where predictor effects are to be modeled, it is often the case that sets of predictors are fixed, and the aim is to choose between fixed model sets. Model selection methods, both Bayesian model selection and Bayesian model averaging, are approaches within the Bayesian paradigm for achieving this aim. In the spatial context, model selection could have a spatial component in the sense that some models may be more appropriate for certain areas of a study region than others. In this work, we examine the use of spatially referenced Bayesian model averaging and Bayesian model selection via a large-scale simulation study accompanied by a small-scale case study. Our results suggest that BMS performs well when a strong regression signature is found.
Keywords: BRugs; Bayesian model averaging; Bayesian model selection; MCMC; R2WinBUGS; spatial.
Conflict of interest statement
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures







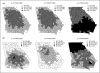
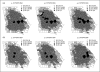

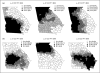


References
-
- Clyde M, Iversen ES. Bayesian model averaging in the M-open framework. In: Damien P, Dellaportas P, Polson NG, Stephens DA, editors. Bayesian theory and applications. Oxford: Oxford University Press; 2015.
-
- Hoeting JA, Madigan D, Raftery AE, et al. Bayesian model averaging: a tutorial. Stat Sci. 1999;14:382–417.
-
- Lesaffre E, Lawson AB. Bayesian biostatistics. 1. West Sussex, UK: Wiley; 2013. p. 534.
-
- Lunn D, Jackson C, Best N, et al. The BUGS book: a practical introduction to bayesian analysis. 1. Boca Raton, FL: CRC Press; 2013. p. 399.
-
- Thomas A, O’hara B, Ligges U, et al. Making BUGS open. R News. 2006;6:12–17.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

