Structural and Molecular Evidence Suggesting Coronavirus-driven Evolution of Mouse Receptor
- PMID: 28035001
- PMCID: PMC5313091
- DOI: 10.1074/jbc.M116.764266
Structural and Molecular Evidence Suggesting Coronavirus-driven Evolution of Mouse Receptor
Abstract
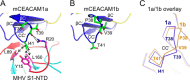
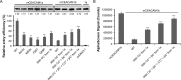
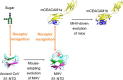
Hosts and pathogens are locked in an evolutionary arms race. To infect mice, mouse hepatitis coronavirus (MHV) has evolved to recognize mouse CEACAM1a (mCEACAM1a) as its receptor. To elude MHV infections, mice may have evolved a variant allele from the Ceacam1a gene, called Ceacam1b, producing mCEACAM1b, which is a much poorer MHV receptor than mCEACAM1a. Previous studies showed that sequence differences between mCEACAM1a and mCEACAM1b in a critical MHV-binding CC' loop partially account for the low receptor activity of mCEACAM1b, but detailed structural and molecular mechanisms for the differential MHV receptor activities of mCEACAM1a and mCEACAM1b remained elusive. Here we have determined the crystal structure of mCEACAM1b and identified the structural differences and additional residue differences between mCEACAM1a and mCEACAM1b that affect MHV binding and entry. These differences include conformational alterations of the CC' loop as well as residue variations in other MHV-binding regions, including β-strands C' and C'' and loop C'C''. Using pseudovirus entry and protein-protein binding assays, we show that substituting the structural and residue features from mCEACAM1b into mCEACAM1a reduced the viral receptor activity of mCEACAM1a, whereas substituting the reverse changes from mCEACAM1a into mCEACAM1b increased the viral receptor activity of mCEACAM1b. These results elucidate the detailed molecular mechanism for how mice may have kept pace in the evolutionary arms race with MHV by undergoing structural and residue changes in the MHV receptor, providing insight into this possible example of pathogen-driven evolution of a host receptor protein.
Keywords: X-ray crystallography; evolution; receptor structure-function; virus; virus entry.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Dawkins R., and Krebs J. R. (1979) Arms races between and within species. Proc. R. Soc. Lond. B Biol. Sci. 205, 489–511 - PubMed
-
- Van Valen L. (1973) A new evolutionary law. Evolutionary Theory 1, 1–30
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

