Effects of DNA supercoiling on chromatin architecture
- PMID: 28035244
- PMCID: PMC5153829
- DOI: 10.1007/s12551-016-0242-6
Effects of DNA supercoiling on chromatin architecture
Abstract
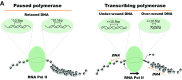
Disruptions in chromatin structure are necessary for the regulation of eukaryotic genomes, from remodelling of nucleosomes at the base pair level through to large-scale chromatin domains that are hundreds of kilobases in size. RNA polymerase is a powerful motor which, prevented from turning with the tight helical pitch of the DNA, generates over-wound DNA ahead of itself and under-wound DNA behind. Mounting evidence supports a central role for transcription-dependent DNA supercoiling in disrupting chromatin structure at all scales. This supercoiling changes the properties of the DNA helix in a manner that substantially alters the binding specificity of DNA binding proteins and complexes, including nucleosomes, polymerases, topoisomerases and transcription factors. For example, transient over-wound DNA destabilises nucleosome core particles ahead of a transcribing polymerase, whereas under-wound DNA facilitates pre-initiation complex formation, transcription factor binding and nucleosome core particle association behind the transcribing polymerase. Importantly, DNA supercoiling can also dissipate through DNA, even in a chromatinised context, to influence both local elements and large chromatin domains. We propose a model in which changes in unconstrained DNA supercoiling influences higher levels of chromatin organisation through the additive effects of DNA supercoiling on both DNA-protein and DNA-nucleosome interactions. This model links small-scale changes in DNA and chromatin to the higher-order fibre and large-scale chromatin structures, providing a mechanism relating gene regulation to chromatin architecture in vivo.
Keywords: DNA supercoiling; Eukaryotic chromatin; Gene regulation; Genome architecture; Protein–DNA.
Conflict of interest statement
Samuel Corless declares that he has no conflict of interest. Nick Gilbert declares that he has no conflict of interest. Ethical approval This article does not contain any studies with human participants or animals performed by any of the authors.
Figures




References
-
- Bates AD, Maxwell A. DNA topology. New York: Oxford University Press; 2005.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

