Robust transcriptional tumor signatures applicable to both formalin-fixed paraffin-embedded and fresh-frozen samples
- PMID: 28036264
- PMCID: PMC5351660
- DOI: 10.18632/oncotarget.14257
Robust transcriptional tumor signatures applicable to both formalin-fixed paraffin-embedded and fresh-frozen samples
Abstract
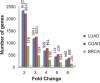
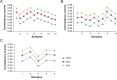
Formalin-fixed paraffin-embedded (FFPE) samples represent a valuable resource for clinical researches. However, FFPE samples are usually considered an unreliable source for gene expression analysis due to the partial RNA degradation. In this study, through comparing gene expression profiles between FFPE samples and paired fresh-frozen (FF) samples for three cancer types, we firstly showed that expression measurements of thousands of genes had at least two-fold change in FFPE samples compared with paired FF samples. Therefore, for a transcriptional signature based on risk scores summarized from the expression levels of the signature genes, the risk score thresholds trained from FFPE (or FF) samples could not be applied to FF (or FFPE) samples. On the other hand, we found that more than 90% of the relative expression orderings (REOs) of gene pairs in the FF samples were maintained in their paired FFPE samples and largely unaffected by the storage time. The result suggested that the REOs of gene pairs were highly robust against partial RNA degradation in FFPE samples. Finally, as a case study, we developed a REOs-based signature to distinguish liver cirrhosis from hepatocellular carcinoma (HCC) using FFPE samples. The signature was validated in four datasets of FFPE samples and eight datasets of FF samples. In conclusion, the valuable FFPE samples can be fully exploited to identify REOs-based diagnostic and prognostic signatures which could be robustly applicable to both FF samples and FFPE samples with degraded RNA.
Keywords: RNA degradation; formalin-fixed paraffin-embedded samples; fresh-frozen samples; gene expression measurements; relative expression orderings.
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures
References
-
- Abdullah-Sayani A, Bueno-de-Mesquita JM, van de Vijver MJ. Technology Insight: tuning into the genetic orchestra using microarrays—limitations of DNA microarrays in clinical practice. Nature clinical practice Oncology. 2006;3:501–516. - PubMed
-
- Blow N. Tissue preparation: Tissue issues. Nature. 2007;448:959–963. - PubMed
-
- Tang W, David FB, Wilson MM, Barwick BG, Leyland-Jones BR, Bouzyk MM. DNA extraction from formalin-fixed, paraffin-embedded tissue. Cold Spring Harbor protocols. 20092009 pdb prot5138. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

