SDH6 and SDH7 Contribute to Anchoring Succinate Dehydrogenase to the Inner Mitochondrial Membrane in Arabidopsis thaliana
- PMID: 28039307
- PMCID: PMC5291046
- DOI: 10.1104/pp.16.01675
SDH6 and SDH7 Contribute to Anchoring Succinate Dehydrogenase to the Inner Mitochondrial Membrane in Arabidopsis thaliana
Abstract
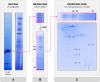
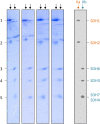
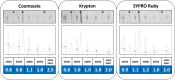
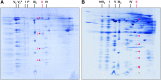
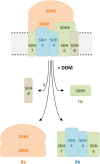
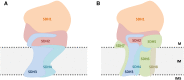
The succinate dehydrogenase complex (complex II) is a highly conserved protein complex composed of the SDH1 to SDH4 subunits in bacteria and in the mitochondria of animals and fungi. The reason for the occurrence of up to four additional subunits in complex II of plants, termed SDH5 to SDH8, so far is a mystery. Here, we present a biochemical approach to investigate the internal subunit arrangement of Arabidopsis (Arabidopsis thaliana) complex II. Using low-concentration detergent treatments, the holo complex is dissected into subcomplexes that are analyzed by a three-dimensional gel electrophoresis system. Protein identifications by mass spectrometry revealed that the largest subcomplex (IIa) represents the succinate dehydrogenase domain composed of SDH1 and SDH2. Another subcomplex (IIb) is composed of the SDH3, SDH4, SDH6, and SDH7 subunits. All four proteins include transmembrane helices and together form the membrane anchor of complex II. Sequence analysis revealed that SDH3 and SDH4 lack helices conserved in other organisms. Using homology modeling and phylogenetic analyses, we present evidence that SDH6 and SDH7 substitute missing sequence stretches of SDH3 and SDH4 in plants. Together with SDH5, which is liberated upon dissection of complex II into subcomplexes, SDH6 and SDH7 also add some hydrophilic mass to plant complex II, which possibly inserts further functions into this smallest protein complex of the oxidative phosphorylation system (which is not so small in plants).
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures


References
-
- Adams KL, Palmer JD (2003) Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol 29: 380–395 - PubMed
-
- Araújo WL, Nunes-Nesi A, Osorio S, Usadel B, Fuentes D, Nagy R, Balbo I, Lehmann M, Studart-Witkowski C, Tohge T, et al. (2011) Antisense inhibition of the iron-sulphur subunit of succinate dehydrogenase enhances photosynthesis and growth in tomato via an organic acid-mediated effect on stomatal aperture. Plant Cell 23: 600–627 - PMC - PubMed
-
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

