Dysregulated long intergenic non-coding RNA modules contribute to heart failure
- PMID: 28040802
- PMCID: PMC5312340
- DOI: 10.18632/oncotarget.10834
Dysregulated long intergenic non-coding RNA modules contribute to heart failure
Abstract
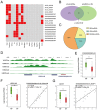
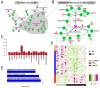
Long intergenic non-coding RNAs (lincRNAs) are emerging as important regulatory molecules involved in diseases including heart failure. However, little is known about how the lincRNAs work together with protein-coding genes (PCGs) contributing to the pathogenesis of heart failure. In this study, we constructed a comprehensive transcriptome profile of lincRNAs, PCGs and miRNAs using RNA-seq and miRNA-seq data of 16 heart failure patients (HFs) and 8 non-failing individuals (NFs). Through integrating lincRNA and PCG expression profiles, we identified HF-associated lincRNA modules. We identified a heart-specific lincRNA module which was significantly enriched for differentially expressed lincRNAs and PCGs. This module was associated with heart failure rather than with other clinical traits such as sex, age, smoking and diabetes mellitus. Moreover, the module was significantly correlated with certain indicators of left ventricular function like ejection fraction and left ventricular end-diastolic diameter, implying the potential of its components as crucial biomarkers. Apart from enhancer-like function, lincRNAs in this module could act as competing endogenous RNAs (ceRNAs) to regulate genes which were associated with left-ventricular systolic function. Our work provided deep insights into the critical roles of lincRNAs in the pathology of heart failure and suggested that they could be valuable biomarkers and therapeutic targets.
Keywords: ceRNA; contraction; heart failure; heart specificity; lincRNAmodule.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8. doi: 10.1126/science.1138341. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

