Quantification of the d-Ala-d-Lac-Terminated Peptidoglycan Structure in Vancomycin-Resistant Enterococcus faecalis Using a Combined Solid-State Nuclear Magnetic Resonance and Mass Spectrometry Analysis
- PMID: 28040891
- PMCID: PMC6906607
- DOI: 10.1021/acs.biochem.6b00774
Quantification of the d-Ala-d-Lac-Terminated Peptidoglycan Structure in Vancomycin-Resistant Enterococcus faecalis Using a Combined Solid-State Nuclear Magnetic Resonance and Mass Spectrometry Analysis
Abstract
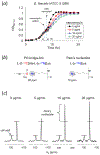
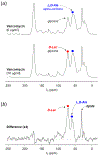
Induction of vancomycin resistance in vancomycin-resistant enterococci (VRE) involves replacement of the d-Ala-d-Ala terminus of peptidoglycan (PG) stems with d-Ala-d-Lac, dramatically reducing the binding affinity of vancomycin for lipid II. Effects from vancomycin resistance induction in Enterococcus faecalis (ATCC 51299) were characterized using a combined solid-state nuclear magnetic resonance (NMR) and liquid chromatography-mass spectrometry (LC-MS) analysis. Solid-state NMR directly measured the total amounts of d-Lac and l,d-Ala metabolized from [2-13C]pyruvate, accumulated Park's nucleotide, and changes to the PG bridge-linking density during the early exponential growth phase (OD660 = 0.4) in intact whole cells of VRE. A high level of accumulation of depsipeptide-substituted Park's nucleotide consistent with the inhibition of the transglycosylation step of PG biosynthesis during the initial phase of vancomycin resistance was observed, while no changes to the PG bridge-linking density following the induction of vancomycin resistance were detected. This indicated that the attachment of the PG bridge to lipid II by the peptidyl transferases was not inhibited by the d-Ala-d-Lac-substituted PG stem structure in VRE. Compositions of mutanolysin-digested isolated cell walls of VRE grown with and without vancomycin resistance induction were determined by LC-MS. Muropeptides with PG stems terminating in d-Ala-d-Lac were found only in VRE grown in the presence of vancomycin. Percentages of muropeptides with a pentapeptide stem terminating in d-Ala-d-Lac for VRE grown in the presence of vancomycin were 26% for the midexponential phase (OD660 = 0.6) and 57% for the stationary growth phase (OD660 = 1.0). These high percentages indicate that d-Ala-d-Lac-substituted lipid II was efficiently utilized for PG biosynthesis in VRE.
Figures



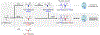
References
-
- Bouhss A, Josseaume N, Allanic D, Crouvoisier M, Gutmann L, Mainardi JL, Mengin-Lecreulx D, van Heijenoort J, and Arthur M (2001) Identification of the UDP-MurNAc-pentapeptide:L-alanine ligase for synthesis of branched peptidoglycan precursors in Enterococcus faecalis, J. Bacteriol 183, 5122–5127. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

