Lipid-Sorting Specificity Encoded in K-Ras Membrane Anchor Regulates Signal Output
- PMID: 28041850
- PMCID: PMC5653213
- DOI: 10.1016/j.cell.2016.11.059
Lipid-Sorting Specificity Encoded in K-Ras Membrane Anchor Regulates Signal Output
Abstract
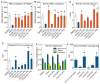
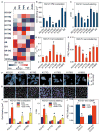
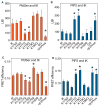
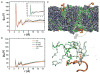
K-Ras is targeted to the plasma membrane by a C-terminal membrane anchor that comprises a farnesyl-cysteine-methyl-ester and a polybasic domain. We used quantitative spatial imaging and atomistic molecular dynamics simulations to examine molecular details of K-Ras plasma membrane binding. We found that the K-Ras anchor binds selected plasma membrane anionic lipids with defined head groups and lipid side chains. The precise amino acid sequence and prenyl group define a combinatorial code for lipid binding that extends beyond simple electrostatics; within this code lysine and arginine residues are non-equivalent and prenyl chain length modifies nascent polybasic domain lipid preferences. The code is realized by distinct dynamic tertiary structures of the anchor on the plasma membrane that govern amino acid side-chain-lipid interactions. An important consequence of this specificity is the ability of such anchors when aggregated to sort subsets of phospholipids into nanoclusters with defined lipid compositions that determine K-Ras signaling output.
Keywords: K-RAS; electron microscopy; molecular dynamics; nanoclustering; phosphatidylserine; polybasic domain.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


References
-
- Bach D, Wachtel E, Borochov N, Senisterra G, Epand RM. Phase behaviour of heteroacid phosphatidylserines and cholesterol. Chem Phys Lipids. 1992;63:105–113.
-
- Barducci A, Bonomi M, Parrinello M. Metadynamics. Wiley Interdiscip Rev Comput Mol Sci. 2011;1:14.
-
- Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, Miura J, Wiener HH, Wright L, Saba SG, et al. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell. 2006;21:481–493. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

