Lipid Desaturation Is a Metabolic Marker and Therapeutic Target of Ovarian Cancer Stem Cells
- PMID: 28041894
- PMCID: PMC5337165
- DOI: 10.1016/j.stem.2016.11.004
Lipid Desaturation Is a Metabolic Marker and Therapeutic Target of Ovarian Cancer Stem Cells
Abstract
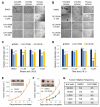
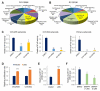
Lack of sensitive single-cell analysis tools has limited the characterization of metabolic activity in cancer stem cells. By hyperspectral-stimulated Raman scattering imaging of single living cells and mass spectrometry analysis of extracted lipids, we report here significantly increased levels of unsaturated lipids in ovarian cancer stem cells (CSCs) as compared to non-CSCs. Higher lipid unsaturation levels were also detected in CSC-enriched spheroids compared to monolayer cultures of ovarian cancer cell lines or primary cells. Inhibition of lipid desaturases effectively eliminated CSCs, suppressed sphere formation in vitro, and blocked tumor initiation capacity in vivo. Mechanistically, we demonstrate that nuclear factor κB (NF-κB) directly regulates the expression levels of lipid desaturases, and inhibition of desaturases blocks NF-κB signaling. Collectively, our findings reveal that increased lipid unsaturation is a metabolic marker for ovarian CSCs and a target for CSC-specific therapy.
Keywords: NF-κB; Raman spectroscopic imaging; SCD1; cancer stem cells; lipid desaturation; ovarian cancer.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures





Comment in
-
Unsaturated Fatty Acids Maintain Cancer Cell Stemness.Cell Stem Cell. 2017 Mar 2;20(3):291-292. doi: 10.1016/j.stem.2017.02.008. Cell Stem Cell. 2017. PMID: 28257705 Free PMC article.
-
Observation-driven inquiry: Raman spectroscopic imaging illuminates cancer lipid metabolism.Stem Cell Investig. 2017 May 26;4:42. doi: 10.21037/sci.2017.04.09. eCollection 2017. Stem Cell Investig. 2017. PMID: 28607916 Free PMC article. No abstract available.
-
Unsaturated fatty acids regulate stemness of ovarian cancer cells through NF-κB.Stem Cell Investig. 2017 Jun 3;4:49. doi: 10.21037/sci.2017.05.07. eCollection 2017. Stem Cell Investig. 2017. PMID: 28607923 Free PMC article. No abstract available.
References
-
- Bjerkvig R, Tysnes BB, Aboody KS, Najbauer J, Terzis AJA. The origin of the cancer stem cell: current controversies and new insights. Nat Rev Cancer. 2005;5:995–995. - PubMed
-
- Camandola S, Leonarduzzi G, Musso T, Varesio L, Carini R, Scavazza A, Chiarpotto E, Baeuerle PA, Poli G. Nuclear factor kB is activated by arachidonic acid but not by eicosapentaenoic acid. Biochem Biophys Res Commun. 1996;229:643–647. - PubMed
-
- Camp CH, Yegnanarayanan S, Eftekhar AA, Sridhar H, Adibi A. Multiplex coherent anti-Stokes Raman scattering (MCARS) for chemicallysensitive, label-free flow cytometry. Opt Express. 2009;17:22879–22889. - PubMed
-
- Camp JCH, Yegnanarayanan S, Eftekhar AA, Adibi A. Label-free flow cytometry using multiplex coherent anti-Stokes Raman scattering (MCARS) for the analysis of biological specimens. Opt Lett. 2011;36:2309–2311. - PubMed
-
- Cheng J-X, Xie XS. Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science. 2015;350:1054. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

