Polarity Reversal by Centrosome Repositioning Primes Cell Scattering during Epithelial-to-Mesenchymal Transition
- PMID: 28041907
- PMCID: PMC5497078
- DOI: 10.1016/j.devcel.2016.12.004
Polarity Reversal by Centrosome Repositioning Primes Cell Scattering during Epithelial-to-Mesenchymal Transition
Abstract
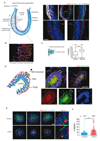
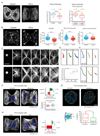
During epithelial-to-mesenchymal transition (EMT), cells lining the tissue periphery break up their cohesion to migrate within the tissue. This dramatic reorganization involves a poorly characterized reorientation of the apicobasal polarity of static epithelial cells into the front-rear polarity of migrating mesenchymal cells. To investigate the spatial coordination of intracellular reorganization with morphological changes, we monitored centrosome positioning during EMT in vivo, in developing mouse embryos and mammary gland, and in vitro, in cultured 3D cell aggregates and micropatterned cell doublets. In all conditions, centrosomes moved from their off-centered position next to intercellular junctions toward extracellular matrix adhesions on the opposite side of the nucleus, resulting in an effective internal polarity reversal. This move appeared to be supported by controlled microtubule network disassembly. Sequential release of cell confinement using dynamic micropatterns, and modulation of microtubule dynamics, confirmed that centrosome repositioning was responsible for further cell disengagement and scattering.
Keywords: EMT; centrosome; cytoskeleton; intercellular junctions; micropatterning; microtubule; migration; polarity; stathmin.
Crown Copyright © 2017. Published by Elsevier Inc. All rights reserved.
Figures





Similar articles
-
Plakins are involved in the regulation of centrosome position in polarized epithelial cells.Biol Cell. 2024 Jul;116(7):e2400048. doi: 10.1111/boc.202400048. Epub 2024 Jun 8. Biol Cell. 2024. PMID: 38850178
-
Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration.Curr Biol. 2009 Jul 14;19(13):1065-74. doi: 10.1016/j.cub.2009.05.065. Epub 2009 Jun 18. Curr Biol. 2009. PMID: 19540120 Free PMC article.
-
Centrosome defines the rear of cells during mesenchymal migration.Mol Biol Cell. 2017 Nov 7;28(23):3240-3251. doi: 10.1091/mbc.E17-06-0366. Epub 2017 Aug 30. Mol Biol Cell. 2017. PMID: 28855377 Free PMC article.
-
Cell polarity in motion: redefining mammary tissue organization through EMT and cell polarity transitions.J Mammary Gland Biol Neoplasia. 2010 Jun;15(2):149-68. doi: 10.1007/s10911-010-9180-2. Epub 2010 May 12. J Mammary Gland Biol Neoplasia. 2010. PMID: 20461450 Review.
-
Cellular dynamics of EMT: lessons from live in vivo imaging of embryonic development.Cell Commun Signal. 2021 Jul 22;19(1):79. doi: 10.1186/s12964-021-00761-8. Cell Commun Signal. 2021. PMID: 34294089 Free PMC article. Review.
Cited by
-
O-GlcNAcylation Regulates Centrosome Behavior and Cell Polarity to Reduce Pulmonary Fibrosis and Maintain the Epithelial Phenotype.Adv Sci (Weinh). 2023 Dec;10(36):e2303545. doi: 10.1002/advs.202303545. Epub 2023 Nov 14. Adv Sci (Weinh). 2023. PMID: 37963851 Free PMC article.
-
The emerging role of microtubules in invasion plasticity.Front Oncol. 2023 Feb 13;13:1118171. doi: 10.3389/fonc.2023.1118171. eCollection 2023. Front Oncol. 2023. PMID: 36860323 Free PMC article. Review.
-
PI3Kδ activity controls plasticity and discriminates between EMT and stemness based on distinct TGFβ signaling.Commun Biol. 2022 Jul 25;5(1):740. doi: 10.1038/s42003-022-03637-w. Commun Biol. 2022. PMID: 35879421 Free PMC article.
-
Paxillin regulates cell polarization and anterograde vesicle trafficking during cell migration.Mol Biol Cell. 2017 Dec 15;28(26):3815-3831. doi: 10.1091/mbc.E17-08-0488. Epub 2017 Oct 18. Mol Biol Cell. 2017. PMID: 29046398 Free PMC article.
-
Regulation of Cell Adhesion and Migration via Microtubule Cytoskeleton Organization, Cell Polarity, and Phosphoinositide Signaling.Biomolecules. 2023 Sep 22;13(10):1430. doi: 10.3390/biom13101430. Biomolecules. 2023. PMID: 37892112 Free PMC article. Review.
References
-
- Akhmanova A, Stehbens SJ, Yap AS. Touch, grasp, deliver and control: functional cross-talk between microtubules and cell adhesions. Traffic. 2009;10:268–274. - PubMed
-
- Anstrom JA, Raff RA. Sea urchin primary mesenchyme cells: Relation of cell polarity to the epithelial-mesenchymal transformation. Dev Biol. 1988;130:57–66. - PubMed
-
- Azioune A, Carpi N, Tseng Q, Théry M, Piel M. Protein micropatterns: A direct printing protocol using deep UVs. Methods Cell Biol. 2010;97:133–146. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

