From structure to redox: The diverse functional roles of disulfides and implications in disease
- PMID: 28044432
- PMCID: PMC5367942
- DOI: 10.1002/pmic.201600391
From structure to redox: The diverse functional roles of disulfides and implications in disease
Abstract
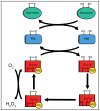
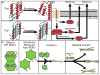
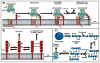
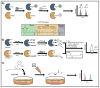
This review provides a comprehensive overview of the functional roles of disulfide bonds and their relevance to human disease. The critical roles of disulfide bonds in protein structure stabilization and redox regulation of protein activity are addressed. Disulfide bonds are essential to the structural stability of many proteins within the secretory pathway and can exist as intramolecular or inter-domain disulfides. The proper formation of these bonds often relies on folding chaperones and oxidases such as members of the protein disulfide isomerase (PDI) family. Many of the PDI family members catalyze disulfide-bond formation, reduction, and isomerization through redox-active disulfides and perturbed PDI activity is characteristic of carcinomas and neurodegenerative diseases. In addition to catalytic function in oxidoreductases, redox-active disulfides are also found on a diverse array of cellular proteins and act to regulate protein activity and localization in response to oxidative changes in the local environment. These redox-active disulfides are either dynamic intramolecular protein disulfides or mixed disulfides with small-molecule thiols generating glutathionylation and cysteinylation adducts. The oxidation and reduction of redox-active disulfides are mediated by cellular reactive oxygen species and activity of reductases, such as glutaredoxin and thioredoxin. Dysregulation of cellular redox conditions and resulting changes in mixed disulfide formation are directly linked to diseases such as cardiovascular disease and Parkinson's disease.
© 2017 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures


References
-
- Sevier CS, Kaiser CA. Formation and transfer of disulphide bonds in living cells. Nat Rev Mol Cell Biol. 2002;3:836–847. - PubMed
-
- Hatahet F, Ruddock LW. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal. 2009;11:2807–2850. - PubMed
-
- Deponte M, Hell K. Disulphide bond formation in the intermembrane space of mitochondria. J Biochem. 2009;146:599–608. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

