Induction of Multiple miR-200/182 Members in the Brains of Mice Are Associated with Acute Herpes Simplex Virus 1 Encephalitis
- PMID: 28045967
- PMCID: PMC5207681
- DOI: 10.1371/journal.pone.0169081
Induction of Multiple miR-200/182 Members in the Brains of Mice Are Associated with Acute Herpes Simplex Virus 1 Encephalitis
Erratum in
-
Correction: Induction of Multiple miR-200/182 Members in the Brains of Mice Are Associated with Acute Herpes Simplex Virus 1 Encephalitis.PLoS One. 2017 Feb 21;12(2):e0172815. doi: 10.1371/journal.pone.0172815. eCollection 2017. PLoS One. 2017. PMID: 28222132 Free PMC article.
Abstract
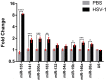
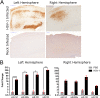
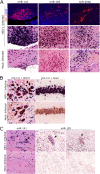
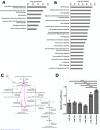
Important roles of microRNAs (miRNAs) in regulating the host response during viral infection have begun to be defined. However, little is known about the functional roles of miRNAs within an in vivo acute viral encephalitis model. We therefore identified global changes in miRNA expression during acute herpes simplex virus type 1 (HSV-1) encephalitis (HSVE) in mice. We found that many of the highly upregulated miRNAs (miR-155, miR-146a and miR-15b) detected in HSV-1 infected brain tissue are known regulators of inflammation and innate immunity. We also observed upregulation of 7 members belonging to the related group of miRNAs, the miR-200 family and miR-182 cluster (miR-200/182). Using in situ hybridization, we found that these miRNAs co-localized to regions of the brain with severe HSVE-related pathology and were upregulated in various cell types including neurons. Induction was apparent but not limited to cells in which HSV-1 was detected by immunohistochemistry, suggesting possible roles of these miRNAs in the host response to viral-induced tissue damage. Bioinformatic prediction combined with gene expression profiling revealed that the induced miR-200/182 members could regulate the biosynthesis of heparan sulfate proteoglycans. Using luciferase assays, we found that miR-96, miR-141, miR-183 and miR-200c all potentially targeted the syndecan-2 gene (Sdc2), which codes for a cell surface heparan sulfate proteoglycan involved in HSV-1 cellular attachment and entry.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Whitley R. New approaches to the therapy of HSV infections. Herpes 2006. August;13(2):53–55. - PubMed
-
- Rozenberg F, Deback C, Agut H. Herpes simplex encephalitis: from virus to therapy. Infect Disord Drug Targets 2011. June;11(3):235–250. - PubMed
-
- Pasieka TJ, Cilloniz C, Carter VS, Rosato P, Katze MG, Leib DA. Functional genomics reveals an essential and specific role for Stat1 in protection of the central nervous system following herpes simplex virus corneal infection. J Virol 2011. December;85(24):12972–12981. 10.1128/JVI.06032-11 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

