High-Throughput Resequencing of Maize Landraces at Genomic Regions Associated with Flowering Time
- PMID: 28045987
- PMCID: PMC5207663
- DOI: 10.1371/journal.pone.0168910
High-Throughput Resequencing of Maize Landraces at Genomic Regions Associated with Flowering Time
Abstract
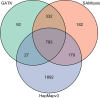
Despite the reduction in the price of sequencing, it remains expensive to sequence and assemble whole, complex genomes of multiple samples for population studies, particularly for large genomes like those of many crop species. Enrichment of target genome regions coupled with next generation sequencing is a cost-effective strategy to obtain sequence information for loci of interest across many individuals, providing a less expensive approach to evaluating sequence variation at the population scale. Here we evaluate amplicon-based enrichment coupled with semiconductor sequencing on a validation set consisting of three maize inbred lines, two hybrids and 19 landrace accessions. We report the use of a multiplexed panel of 319 PCR assays that target 20 candidate loci associated with photoperiod sensitivity in maize while requiring 25 ng or less of starting DNA per sample. Enriched regions had an average on-target sequence read depth of 105 with 98% of the sequence data mapping to the maize 'B73' reference and 80% of the reads mapping to the target interval. Sequence reads were aligned to B73 and 1,486 and 1,244 variants were called using SAMtools and GATK, respectively. Of the variants called by both SAMtools and GATK, 30% were not previously reported in maize. Due to the high sequence read depth, heterozygote genotypes could be called with at least 92.5% accuracy in hybrid materials using GATK. The genetic data are congruent with previous reports of high total genetic diversity and substantial population differentiation among maize landraces. In conclusion, semiconductor sequencing of highly multiplexed PCR reactions is a cost-effective strategy for resequencing targeted genomic loci in diverse maize materials.
Conflict of interest statement
One author is currently employed by Monsanto Company. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures




References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

