Proximity Interactions among Basal Body Components in Trypanosoma brucei Identify Novel Regulators of Basal Body Biogenesis and Inheritance
- PMID: 28049148
- PMCID: PMC5210500
- DOI: 10.1128/mBio.02120-16
Proximity Interactions among Basal Body Components in Trypanosoma brucei Identify Novel Regulators of Basal Body Biogenesis and Inheritance
Abstract
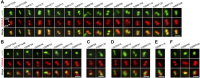
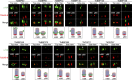
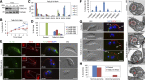
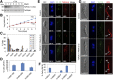
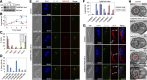
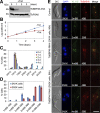
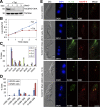
The basal body shares similar architecture with centrioles in animals and is involved in nucleating flagellar axonemal microtubules in flagellated eukaryotes. The early-branching Trypanosoma brucei possesses a motile flagellum nucleated from the basal body that consists of a mature basal body and an adjacent pro-basal body. Little is known about the basal body proteome and its roles in basal body biogenesis and flagellar axoneme assembly in T. brucei Here, we report the identification of 14 conserved centriole/basal body protein homologs and 25 trypanosome-specific basal body proteins. These proteins localize to distinct subdomains of the basal body, and several of them form a ring-like structure surrounding the basal body barrel. Functional characterization of representative basal body proteins revealed distinct roles in basal body duplication/separation and flagellar axoneme assembly. Overall, this work identified novel proteins required for basal body duplication and separation and uncovered new functions of conserved basal body proteins in basal body duplication and separation, highlighting an unusual mechanism of basal body biogenesis and inheritance in this early divergent eukaryote.
Importance: The basal body in the early-branching protozoan Trypanosoma brucei nucleates flagellum assembly and also regulates organelle segregation, cell morphogenesis, and cell division. However, the molecular composition and the assembly process of the basal body remain poorly understood. Here, we identify 14 conserved basal body proteins and 25 trypanosome-specific basal body proteins via bioinformatics, localization-based screening, and proximity-dependent biotin identification. We further localized these proteins to distinct subdomains of the basal body by using fluorescence microscopy and superresolution microscopy, discovered novel regulators of basal body duplication and separation, and uncovered new functions of conserved basal body proteins in basal body duplication and separation. This work lays the foundation for dissecting the mechanisms underlying basal body biogenesis and inheritance in T. brucei.
Copyright © 2017 Dang et al.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
