CRISPR-Cas9: a promising tool for gene editing on induced pluripotent stem cells
- PMID: 28049282
- PMCID: PMC5214730
- DOI: 10.3904/kjim.2016.198
CRISPR-Cas9: a promising tool for gene editing on induced pluripotent stem cells
Abstract
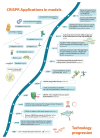
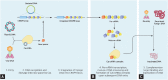
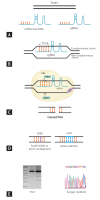
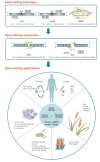
Recent advances in genome editing with programmable nucleases have opened up new avenues for multiple applications, from basic research to clinical therapy. The ease of use of the technology-and particularly clustered regularly interspaced short palindromic repeats (CRISPR)-will allow us to improve our understanding of genomic variation in disease processes via cellular and animal models. Here, we highlight the progress made in correcting gene mutations in monogenic hereditary disorders and discuss various CRISPR-associated applications, such as cancer research, synthetic biology, and gene therapy using induced pluripotent stem cells. The challenges, ethical issues, and future prospects of CRISPR-based systems for human research are also discussed.
Keywords: Clustered regularly interspaced short palindromic repeats; Clustered regularly interspaced short palindromic repeats-Cas9; Gene editing; Genetic therapy; Induced pluripotent stem cells.
Conflict of interest statement
No potential conflict of interest relevant to this article was reported.
Figures
References
-
- Lander ES. Cutting the Gordian helix: regulating genomic testing in the era of precision medicine. N Engl J Med. 2015;372:1185–1186. - PubMed
-
- Jameson JL, Longo DL. Precision medicine: personalized, problematic, and promising. N Engl J Med. 2015;372:2229–2234. - PubMed
-
- Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
