Synthetic DNA Synthesis and Assembly: Putting the Synthetic in Synthetic Biology
- PMID: 28049645
- PMCID: PMC5204324
- DOI: 10.1101/cshperspect.a023812
Synthetic DNA Synthesis and Assembly: Putting the Synthetic in Synthetic Biology
Abstract
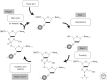
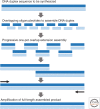
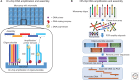
The chemical synthesis of DNA oligonucleotides and their assembly into synthons, genes, circuits, and even entire genomes by gene synthesis methods has become an enabling technology for modern molecular biology and enables the design, build, test, learn, and repeat cycle underpinning innovations in synthetic biology. In this perspective, we briefly review the techniques and technologies that enable the synthesis of DNA oligonucleotides and their assembly into larger DNA constructs with a focus on recent advancements that have sought to reduce synthesis cost and increase sequence fidelity. The development of lower-cost methods to produce high-quality synthetic DNA will allow for the exploration of larger biological hypotheses by lowering the cost of use and help to close the DNA read-write cost gap.
Copyright © 2017 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures


References
-
- Agarwal KL, Buchi H, Caruthers MH, Gupta N, Khorana HG, Kleppe K, Kumar A, Ohtsuka E, Rajbhandary UL, Van de Sande JH, et al. 1970. Total synthesis of the gene for an alanine transfer ribonucleic acid from yeast. Nature 227: 27–34. - PubMed
-
- Au LC, Yang FY, Yang WJ, Lo SH, Kao CF. 1998. Gene synthesis by an LCR-based approach: High-level production of leptin-l54 using synthetic gene in Escherichia coli. Biochem Biophys Res Commun 248: 200–203. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
