Gpr132 sensing of lactate mediates tumor-macrophage interplay to promote breast cancer metastasis
- PMID: 28049847
- PMCID: PMC5255630
- DOI: 10.1073/pnas.1614035114
Gpr132 sensing of lactate mediates tumor-macrophage interplay to promote breast cancer metastasis
Abstract
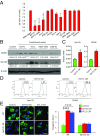
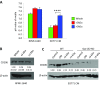
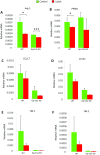
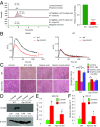
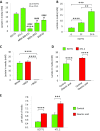
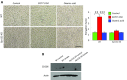
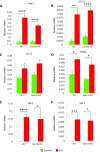
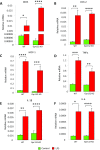
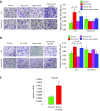
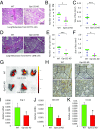
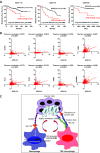
Macrophages are prominent immune cells in the tumor microenvironment that exert potent effects on cancer metastasis. However, the signals and receivers for the tumor-macrophage communication remain enigmatic. Here, we show that G protein-coupled receptor 132 (Gpr132) functions as a key macrophage sensor of the rising lactate in the acidic tumor milieu to mediate the reciprocal interaction between cancer cells and macrophages during breast cancer metastasis. Lactate activates macrophage Gpr132 to promote the alternatively activated macrophage (M2)-like phenotype, which, in turn, facilitates cancer cell adhesion, migration, and invasion. Consequently, Gpr132 deletion reduces M2 macrophages and impedes breast cancer lung metastasis in mice. Clinically, Gpr132 expression positively correlates with M2 macrophages, metastasis, and poor prognosis in patients with breast cancer. These findings uncover the lactate-Gpr132 axis as a driver of breast cancer metastasis by stimulating tumor-macrophage interplay, and reveal potential new therapeutic targets for breast cancer treatment.
Keywords: Gpr132; breast cancer; lactate; macrophage; metastasis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

