Co-Expression Analysis of Blood Cell Genome Expression to Preliminary Investigation of Regulatory Mechanisms in Uremia
- PMID: 28050009
- PMCID: PMC5228761
- DOI: 10.12659/msm.899385
Co-Expression Analysis of Blood Cell Genome Expression to Preliminary Investigation of Regulatory Mechanisms in Uremia
Abstract
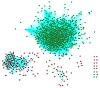
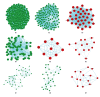
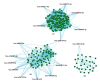
BACKGROUND Uremia involves a series of clinical manifestations and is a common syndrome that occurs in nearly all end-stage kidney diseases. However, the exact genetic and/or molecular mechanisms that underlie uremia remain poorly understood. MATERIAL AND METHODS In this case-control study, we analyzed whole-genome microarray of 75 uremia patients and 20 healthy controls to investigate changes in gene expression and cellular mechanisms relevant to uremia. Gene co-expression network analysis was performed to construct co-expression networks using differentially expressed genes (DEGs) in uremia. We then determined hub models of co-expressed gene networks by MCODE, and we used miRNA enrichment analysis to detect key miRNAs in each hub module. RESULTS We found nine co-expressed hub modules implicated in uremia. These modules were enriched in specific biological functions, including "proteolysis", "membrane-enclosed lumen", and "apoptosis". Finally, miRNA enrichment analysis to detect key miRNAs in each hub module found 15 miRNAs that were specifically targeted to uremia-related hub modules. Of these, miRNA-21-3p and miRNA-210-3p have been identified in other studies as being important for uremia. CONCLUSIONS In summary, our study connected biological functions, genes, and miRNAs that underpin the network modules that can be used to elucidate the molecular mechanisms involved in uremia.
Figures
References
-
- Schreiner G, Maher J. Uremia. Springfield, IL: Charles C. Thomas; 1960. Biochemistry of uremia; pp. 55–85.
-
- Berman N. End-of-life matters in chronic renal failure. Curr Opin Support Palliat Care. 2014;8(4):371–77. - PubMed
-
- Almeras C, Argilés A. The general picture of uremia. Semin Dial. 2009;22(4):329–33. - PubMed
-
- Chikotas N, Gunderman A, Oman T. Uremic syndrome and end-stage renal disease: Physical manifestations and beyond. J Am Acad Nurse Pract. 2006;18(5):195–202. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

