High Prevalences and a Wide Genetic Diversity of Simian Retroviruses in Non-human Primate Bushmeat in Rural Areas of the Democratic Republic of Congo
- PMID: 28050688
- PMCID: PMC5360875
- DOI: 10.1007/s10393-016-1202-0
High Prevalences and a Wide Genetic Diversity of Simian Retroviruses in Non-human Primate Bushmeat in Rural Areas of the Democratic Republic of Congo
Erratum in
-
Erratum to: High Prevalences and a Wide Genetic Diversity of Simian Retroviruses in Non-human Primate Bushmeat in Rural Areas of the Democratic Republic of Congo.Ecohealth. 2017 Mar;14(1):115. doi: 10.1007/s10393-017-1223-3. Ecohealth. 2017. PMID: 28258524 No abstract available.
Abstract
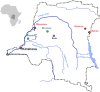
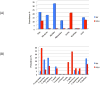
Like the majority of emerging infectious diseases, HIV and HTLV are of zoonotic origin. Here we assess the risk of cross-species transmissions of their simian counterparts, SIV and STLV, from non-human primates (NHP) to humans in the Democratic Republic of Congo (DRC). A total of 331 samples, derived from NHP bushmeat, were collected as dried blood spots (DBS, n = 283) or as tissue samples (n = 36) at remote forest sites mainly in northern and eastern DRC. SIV antibody prevalences in DBS were estimated with a novel high throughput immunoassay with antigens representing the actual known diversity of HIV/SIV lineages. Antibody-positive samples were confirmed by PCR and sequence analysis. Screening for STLV infection was done with universal primers in tax, and new strains were further characterized in LTR. SIV and STLV infection in tissue samples was done by PCR only. Overall, 5 and 15.4% of NHP bushmeat was infected with SIV and STLV, respectively. A new SIV lineage was identified in Allen's swamp monkeys (Allenopithecus nigroviridis). Three new STLV-1 subtypes were identified in Allen's swamp monkeys (Allenopithecus nigroviridis), blue monkeys (Cercopithecus mitis), red-tailed guenons (Cercopithecus ascanius schmidti) and agile mangabeys (Cercocebus agilis). SIV and STLV prevalences varied according to species and geographic region. Our study illustrates clearly, even on a small sample size from a limited number of geographic areas, that our knowledge on the genetic diversity and geographic distribution of simian retroviruses is still limited and that humans continue to be exposed to relative high proportions on infected NHP bushmeat.
Keywords: Bushmeat; Democratic Republic of Congo; Non-human primate; Retrovirus; SIV; STLV.
Conflict of interest statement
Figures


References
-
- Aghokeng AF, Ayouba A, Mpoudi-Ngole E, Loul S, Liegeois F, Delaporte E, Peeters M. Extensive survey on the prevalence and genetic diversity of SIVs in primate bushmeat provides insights into risks for potential new cross-species transmissions. Infect Genet Evol. 2010;201010:386–396. doi: 10.1016/j.meegid.2009.04.014. - DOI - PMC - PubMed
-
- Ahuka-Mundeke S, Liegeois F, Ayouba A, Foupouapouognini Y, Nerrienet E, Delaporte E, Peeters M. Full-length genome sequence of a simian immunodeficiency virus (SIV) infecting a captive agile mangabey (Cercocebus agilis) is closely related to SIVrcm infecting wild red-capped mangabeys (Cercocebus torquatus) in Cameroon. J Gen Virol. 2010;91:2959–64. doi: 10.1099/vir.0.025767-0. - DOI - PMC - PubMed
-
- Ahuka-Mundeke S, Ayouba A, Mbala-Kingebeni P, Liegeois F, Esteban A, Lunguya-Metila O, Demba D, Bilulu G, Mbenzo-Abokome V, Inogwabini BI, Muyembe-Tamfum JJ, Delaporte E, Peeters M. Novel multiplexed HIV/simian immunodeficiency virus antibody detection assay. Emerg Infect Dis. 2011;17:2277–86. doi: 10.3201/eid1712.110783. - DOI - PMC - PubMed
-
- Ahuka-Mundeke S, Mbala-Kingebeni P, Liegeois F, Ayouba A, Lunguya-Metila O, Demba D, Bilulu G, Mbenzo-Abokome V, Inogwabini BI, Muyembe-Tamfum JJ, Delaporte E, Peeters M. Identification and molecular characterization of new simian T cell lymphotropic viruses in nonhuman primates bushmeat from the Democratic Republic of Congo. AIDS Res Hum Retroviruses. 2012;28:628–35. doi: 10.1089/AID.2011.0211. - DOI - PMC - PubMed
-
- Ahuka-Mundeke S, Lunguya-Metila O, Mbenzo-Abokome V, Butel C, Inogwabini B, Omasombo V, Muyembe-Tamfum JJ, Georgiev AV, Muller MN, Ndjango JB, Li Y, Delaporte E, Hahn BH, Peeters M, Ayouba A. Genetic diversity of STLV-2 and interspecies transmission of STLV-3 in wild-living bonobos. Virus evolution. 2016 in press. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

