Mechanisms of Yersinia YopO kinase substrate specificity
- PMID: 28051168
- PMCID: PMC5209680
- DOI: 10.1038/srep39998
Mechanisms of Yersinia YopO kinase substrate specificity
Abstract
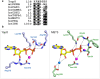
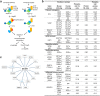
Yersinia bacteria cause a range of human diseases, including yersiniosis, Far East scarlet-like fever and the plague. Yersiniae modulate and evade host immune defences through injection of Yersinia outer proteins (Yops) into phagocytic cells. One of the Yops, YopO (also known as YpkA) obstructs phagocytosis through disrupting actin filament regulation processes - inhibiting polymerization-promoting signaling through sequestration of Rac/Rho family GTPases and by using monomeric actin as bait to recruit and phosphorylate host actin-regulating proteins. Here we set out to identify mechanisms of specificity in protein phosphorylation by YopO that would clarify its effects on cytoskeleton disruption. We report the MgADP structure of Yersinia enterocolitica YopO in complex with actin, which reveals its active site architecture. Using a proteome-wide kinase-interacting substrate screening (KISS) method, we identified that YopO phosphorylates a wide range of actin-modulating proteins and located their phosphorylation sites by mass spectrometry. Using artificial substrates we clarified YopO's substrate length requirements and its phosphorylation consensus sequence. These findings provide fresh insight into the mechanism of the YopO kinase and demonstrate that YopO executes a specific strategy targeting actin-modulating proteins, across multiple functionalities, to compete for control of their native phospho-signaling, thus hampering the cytoskeletal processes required for macrophage phagocytosis.
Figures




References
-
- Viboud G. I. & Bliska J. B. Yersinia outer proteins: Role in Modulation of Host Cell Signaling Responses and Pathogenesis. Annu Rev Microb 59, 69–89 (2005). - PubMed
-
- Hamid N. et al.. YopH dephosphorylates Cas and Fyn-binding protein in macrophages. Microb Pathog 27, 231–242 (1999). - PubMed
-
- Black D. S. & Bliska J. B. The RhoGAP activity of the Yersinia pseudotuberculosis cytotoxin YopE is required for antiphagocytic function and virulence. Mol Microbiol 37, 515–527 (2000). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

