HIV-1 Env associates with HLA-C free-chains at the cell membrane modulating viral infectivity
- PMID: 28051183
- PMCID: PMC5209703
- DOI: 10.1038/srep40037
HIV-1 Env associates with HLA-C free-chains at the cell membrane modulating viral infectivity
Erratum in
-
Corrigendum: HIV-1 Env associates with HLA-C free-chains at the cell membrane modulating viral infectivity.Sci Rep. 2018 May 22;8:46991. doi: 10.1038/srep46991. Sci Rep. 2018. PMID: 29786690 Free PMC article.
Abstract
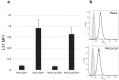
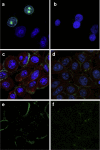
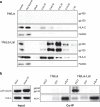
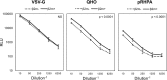
HLA-C has been demonstrated to associate with HIV-1 envelope glycoprotein (Env). Virions lacking HLA-C have reduced infectivity and increased susceptibility to neutralizing antibodies. Like all others MHC-I molecules, HLA-C requires β2-microglobulin (β2m) for appropriate folding and expression on the cell membrane but this association is weaker, thus generating HLA-C free-chains on the cell surface. In this study, we deepen the understanding of HLA-C and Env association by showing that HIV-1 specifically increases the amount of HLA-C free chains, not bound to β2m, on the membrane of infected cells. The association between Env and HLA-C takes place at the cell membrane requiring β2m to occur. We report that the enhanced infectivity conferred to HIV-1 by HLA-C specifically involves HLA-C free chain molecules that have been correctly assembled with β2m. HIV-1 Env-pseudotyped viruses produced in the absence of β2m are less infectious than those produced in the presence of β2m. We hypothesize that the conformation and surface expression of HLA-C molecules could be a discriminant for the association with Env. Binding stability to β2m may confer to HLA-C the ability to preferentially act either as a conventional immune-competent molecule or as an accessory molecule involved in HIV-1 infectivity.
Figures
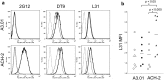
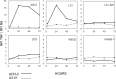
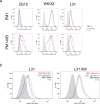
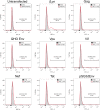
Similar articles
-
Stability and Expression Levels of HLA-C on the Cell Membrane Modulate HIV-1 Infectivity.J Virol. 2017 Dec 14;92(1):e01711-17. doi: 10.1128/JVI.01711-17. Print 2018 Jan 1. J Virol. 2017. PMID: 29070683 Free PMC article.
-
Glycosyl-Phosphatidylinositol-Anchored Anti-HIV Env Single-Chain Variable Fragments Interfere with HIV-1 Env Processing and Viral Infectivity.J Virol. 2018 Mar 14;92(7):e02080-17. doi: 10.1128/JVI.02080-17. Print 2018 Apr 1. J Virol. 2018. PMID: 29321330 Free PMC article.
-
Membrane expression of HLA-Cw4 free chains in activated T cells of transgenic mice.Immunogenetics. 1995;42(5):368-75. doi: 10.1007/BF00179398. Immunogenetics. 1995. PMID: 7590970
-
SERINC as a Restriction Factor to Inhibit Viral Infectivity and the Interaction with HIV.J Immunol Res. 2017;2017:1548905. doi: 10.1155/2017/1548905. Epub 2017 Nov 22. J Immunol Res. 2017. PMID: 29359168 Free PMC article. Review.
-
Four Faces of Cell-Surface HLA Class-I: Their Antigenic and Immunogenic Divergence Generating Novel Targets for Vaccines.Vaccines (Basel). 2022 Feb 21;10(2):339. doi: 10.3390/vaccines10020339. Vaccines (Basel). 2022. PMID: 35214796 Free PMC article. Review.
Cited by
-
The Role of Beta2-Microglobulin in Central Nervous System Disease.Cell Mol Neurobiol. 2024 May 14;44(1):46. doi: 10.1007/s10571-024-01481-6. Cell Mol Neurobiol. 2024. PMID: 38743119 Free PMC article. Review.
-
Molecular Signatures of a TLR4 Agonist-Adjuvanted HIV-1 Vaccine Candidate in Humans.Front Immunol. 2018 Feb 26;9:301. doi: 10.3389/fimmu.2018.00301. eCollection 2018. Front Immunol. 2018. PMID: 29535712 Free PMC article.
-
Modulation of Human Leukocyte Antigen-C by Human Cytomegalovirus Stimulates KIR2DS1 Recognition by Natural Killer Cells.Front Immunol. 2017 Mar 29;8:298. doi: 10.3389/fimmu.2017.00298. eCollection 2017. Front Immunol. 2017. PMID: 28424684 Free PMC article.
-
HIV-1-Associated Neurocognitive Disorders: Is HLA-C Binding Stability to β2-Microglobulin a Missing Piece of the Pathogenetic Puzzle?Front Neurol. 2018 Sep 21;9:791. doi: 10.3389/fneur.2018.00791. eCollection 2018. Front Neurol. 2018. PMID: 30298049 Free PMC article.
-
Contribution of the HIV-1 Envelope Glycoprotein to AIDS Pathogenesis and Clinical Progression.Biomedicines. 2022 Sep 2;10(9):2172. doi: 10.3390/biomedicines10092172. Biomedicines. 2022. PMID: 36140273 Free PMC article. Review.
References
-
- Esser M. T. et al. Differential incorporation of CD45, CD80 (B7-1), CD86 (B7-2), and major histocompatibility complex class I and II molecules into human immunodeficiency virus type 1 virions and microvesicles: implications for viral pathogenesis and immune regulation. J Virol 75, 6173–6182 (2001). - PMC - PubMed
-
- Cosma A. et al. Enhanced HIV infectivity and changes in GP120 conformation associated with viral incorporation of human leucocyte antigen class I molecules. Aids 13, 2033–2042 (1999). - PubMed
-
- Baroni M. et al. HLA-C is necessary for optimal human immunodeficiency virus type 1 infection of human peripheral blood CD4 lymphocytes. The Journal of general virology 91, 235–241 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

