Mapping Polyclonal HIV-1 Antibody Responses via Next-Generation Neutralization Fingerprinting
- PMID: 28052137
- PMCID: PMC5241146
- DOI: 10.1371/journal.ppat.1006148
Mapping Polyclonal HIV-1 Antibody Responses via Next-Generation Neutralization Fingerprinting
Abstract
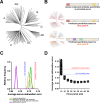
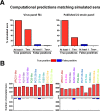
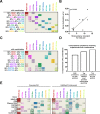
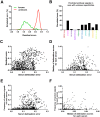
Computational neutralization fingerprinting, NFP, is an efficient and accurate method for predicting the epitope specificities of polyclonal antibody responses to HIV-1 infection. Here, we present next-generation NFP algorithms that substantially improve prediction accuracy for individual donors and enable serologic analysis for entire cohorts. Specifically, we developed algorithms for: (a) selection of optimized virus neutralization panels for NFP analysis, (b) estimation of NFP prediction confidence for each serum sample, and (c) identification of sera with potentially novel epitope specificities. At the individual donor level, the next-generation NFP algorithms particularly improved the ability to detect multiple epitope specificities in a sample, as confirmed both for computationally simulated polyclonal sera and for samples from HIV-infected donors. Specifically, the next-generation NFP algorithms detected multiple specificities in twice as many samples of simulated sera. Further, unlike the first-generation NFP, the new algorithms were able to detect both of the previously confirmed antibody specificities, VRC01-like and PG9-like, in donor CHAVI 0219. At the cohort level, analysis of ~150 broadly neutralizing HIV-infected donor samples suggested a potential connection between clade of infection and types of elicited epitope specificities. Most notably, while 10E8-like antibodies were observed in infections from different clades, an enrichment of such antibodies was predicted for clade B samples. Ultimately, such large-scale analyses of antibody responses to HIV-1 infection can help guide the design of epitope-specific vaccines that are tailored to take into account the prevalence of infecting clades within a specific geographic region. Overall, the next-generation NFP technology will be an important tool for the analysis of broadly neutralizing polyclonal antibody responses against HIV-1.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Korber B, Gaschen B, Yusim K, Thakallapally R, Kesmir C, Detours V. Evolutionary and immunological implications of contemporary HIV-1 variation. Br Med Bull. 2001;58:19–42. Epub 2001/11/21. - PubMed
-
- Karlsson Hedestam GB, Fouchier RAM, Phogat S, Burton DR, Sodroski J, Wyatt RT. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat Rev Micro. 2008;6(2):143–55. - PubMed
-
- Corti D, Langedijk JP, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PloS one. 2010;5(1):e8805 10.1371/journal.pone.0008805 - DOI - PMC - PubMed

