FEELnc: a tool for long non-coding RNA annotation and its application to the dog transcriptome
- PMID: 28053114
- PMCID: PMC5416892
- DOI: 10.1093/nar/gkw1306
FEELnc: a tool for long non-coding RNA annotation and its application to the dog transcriptome
Abstract
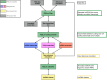
Whole transcriptome sequencing (RNA-seq) has become a standard for cataloguing and monitoring RNA populations. One of the main bottlenecks, however, is to correctly identify the different classes of RNAs among the plethora of reconstructed transcripts, particularly those that will be translated (mRNAs) from the class of long non-coding RNAs (lncRNAs). Here, we present FEELnc (FlExible Extraction of LncRNAs), an alignment-free program that accurately annotates lncRNAs based on a Random Forest model trained with general features such as multi k-mer frequencies and relaxed open reading frames. Benchmarking versus five state-of-the-art tools shows that FEELnc achieves similar or better classification performance on GENCODE and NONCODE data sets. The program also provides specific modules that enable the user to fine-tune classification accuracy, to formalize the annotation of lncRNA classes and to identify lncRNAs even in the absence of a training set of non-coding RNAs. We used FEELnc on a real data set comprising 20 canine RNA-seq samples produced by the European LUPA consortium to substantially expand the canine genome annotation to include 10 374 novel lncRNAs and 58 640 mRNA transcripts. FEELnc moves beyond conventional coding potential classifiers by providing a standardized and complete solution for annotating lncRNAs and is freely available at https://github.com/tderrien/FEELnc.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme.BMC Bioinformatics. 2014 Sep 19;15(1):311. doi: 10.1186/1471-2105-15-311. BMC Bioinformatics. 2014. PMID: 25239089 Free PMC article.
-
A systematic evaluation of bioinformatics tools for identification of long noncoding RNAs.RNA. 2021 Jan;27(1):80-98. doi: 10.1261/rna.074724.120. Epub 2020 Oct 14. RNA. 2021. PMID: 33055239 Free PMC article.
-
Long noncoding RNA repertoire in chicken liver and adipose tissue.Genet Sel Evol. 2017 Jan 10;49(1):6. doi: 10.1186/s12711-016-0275-0. Genet Sel Evol. 2017. PMID: 28073357 Free PMC article.
-
Multi-omics annotation of human long non-coding RNAs.Biochem Soc Trans. 2020 Aug 28;48(4):1545-1556. doi: 10.1042/BST20191063. Biochem Soc Trans. 2020. PMID: 32756901 Review.
-
Strategies to Annotate and Characterize Long Noncoding RNAs: Advantages and Pitfalls.Trends Genet. 2018 Sep;34(9):704-721. doi: 10.1016/j.tig.2018.06.002. Epub 2018 Jul 17. Trends Genet. 2018. PMID: 30017313 Review.
Cited by
-
circRNA-TBC1D4, circRNA-NAALAD2 and circRNA-TGFBR3: Selected Key circRNAs in Neuroblastoma and Their Associations with Clinical Features.Cancer Manag Res. 2021 May 28;13:4271-4281. doi: 10.2147/CMAR.S297316. eCollection 2021. Cancer Manag Res. 2021. PMID: 34093041 Free PMC article.
-
Identification and functional prediction of long non-coding RNAs of rice (Oryza sativa L.) at reproductive stage under salinity stress.Mol Biol Rep. 2021 Mar;48(3):2261-2271. doi: 10.1007/s11033-021-06246-8. Epub 2021 Mar 19. Mol Biol Rep. 2021. PMID: 33742326
-
The profiles and clinical significance of extraocular muscle-expressed lncRNAs and mRNAs in oculomotor nerve palsy.Front Mol Neurosci. 2023 Dec 20;16:1293344. doi: 10.3389/fnmol.2023.1293344. eCollection 2023. Front Mol Neurosci. 2023. PMID: 38173464 Free PMC article.
-
Transcriptome analysis reveals the potential roles of long non-coding RNAs in feed efficiency of chicken.Sci Rep. 2022 Feb 15;12(1):2558. doi: 10.1038/s41598-022-06528-6. Sci Rep. 2022. PMID: 35169237 Free PMC article.
-
Pulmonary artery embolism: comprehensive transcriptomic analysis in understanding the pathogenic mechanisms of the disease.BMC Genomics. 2023 Jan 9;24(1):10. doi: 10.1186/s12864-023-09110-0. BMC Genomics. 2023. PMID: 36624378 Free PMC article.
References
-
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C. et al. . The transcriptional landscape of the mammalian genome. Science. 2005; 309:1559–1563. - PubMed
-
- Legeai F., Derrien T.. Identification of long non-coding RNAs in insects genomes. Curr. Opin. Insect Sci. 2015; 7:37–44. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

