A 'Semi-Protected Oligonucleotide Recombination' Assay for DNA Mismatch Repair in vivo Suggests Different Modes of Repair for Lagging Strand Mismatches
- PMID: 28053122
- PMCID: PMC5416779
- DOI: 10.1093/nar/gkw1339
A 'Semi-Protected Oligonucleotide Recombination' Assay for DNA Mismatch Repair in vivo Suggests Different Modes of Repair for Lagging Strand Mismatches
Abstract
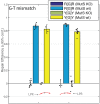
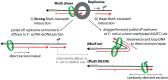
In Escherichia coli, a DNA mismatch repair (MMR) pathway corrects errors that occur during DNA replication by coordinating the excision and re-synthesis of a long tract of the newly-replicated DNA between an epigenetic signal (a hemi-methylated d(GATC) site or a single-stranded nick) and the replication error after the error is identified by protein MutS. Recent observations suggest that this 'long-patch repair' between these sites is coordinated in the same direction of replication by the replisome. Here, we have developed a new assay that uniquely allows us to introduce targeted 'mismatches' directly into the replication fork via oligonucleotide recombination, examine the directionality of MMR, and quantify the nucleotide-dependence, sequence context-dependence, and strand-dependence of their repair in vivo-something otherwise nearly impossible to achieve. We find that repair of genomic lagging strand mismatches occurs bi-directionally in E. coli and that, while all MutS-recognized mismatches had been thought to be repaired in a consistent manner, the directional bias of repair and the effects of mutations in MutS are dependent on the molecular species of the mismatch. Because oligonucleotide recombination is routinely performed in both prokaryotic and eukaryotic cells, we expect this assay will be broadly applicable for investigating mechanisms of MMR in vivo.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


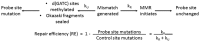
Similar articles
-
Endonuclease-independent DNA mismatch repair processes on the lagging strand.DNA Repair (Amst). 2018 Aug;68:41-49. doi: 10.1016/j.dnarep.2018.06.002. Epub 2018 Jun 12. DNA Repair (Amst). 2018. PMID: 29929046 Free PMC article.
-
Identification of factors influencing strand bias in oligonucleotide-mediated recombination in Escherichia coli.Nucleic Acids Res. 2003 Nov 15;31(22):6674-87. doi: 10.1093/nar/gkg844. Nucleic Acids Res. 2003. PMID: 14602928 Free PMC article.
-
Oligonucleotide transformation of yeast reveals mismatch repair complexes to be differentially active on DNA replication strands.Proc Natl Acad Sci U S A. 2007 Jul 3;104(27):11352-7. doi: 10.1073/pnas.0704695104. Epub 2007 Jun 25. Proc Natl Acad Sci U S A. 2007. PMID: 17592146 Free PMC article.
-
MutL: conducting the cell's response to mismatched and misaligned DNA.Bioessays. 2010 Jan;32(1):51-9. doi: 10.1002/bies.200900089. Bioessays. 2010. PMID: 19953589 Review.
-
DNA mismatch repair.Annu Rev Biochem. 2005;74:681-710. doi: 10.1146/annurev.biochem.74.082803.133243. Annu Rev Biochem. 2005. PMID: 15952900 Review.
Cited by
-
Genome editing outcomes reveal mycobacterial NucS participates in a short-patch repair of DNA mismatches.Nucleic Acids Res. 2024 Nov 11;52(20):12295-12307. doi: 10.1093/nar/gkae402. Nucleic Acids Res. 2024. PMID: 38747340 Free PMC article.
-
Single gold-bridged nanoprobes for identification of single point DNA mutations.Nat Commun. 2019 Feb 19;10(1):836. doi: 10.1038/s41467-019-08769-y. Nat Commun. 2019. PMID: 30783107 Free PMC article.
-
Endonuclease-independent DNA mismatch repair processes on the lagging strand.DNA Repair (Amst). 2018 Aug;68:41-49. doi: 10.1016/j.dnarep.2018.06.002. Epub 2018 Jun 12. DNA Repair (Amst). 2018. PMID: 29929046 Free PMC article.
-
Genome Editing Outcomes Reveal Mycobacterial NucS Participates in a Short-Patch Repair of DNA Mismatches.bioRxiv [Preprint]. 2023 Oct 23:2023.10.23.563644. doi: 10.1101/2023.10.23.563644. bioRxiv. 2023. Update in: Nucleic Acids Res. 2024 Nov 11;52(20):12295-12307. doi: 10.1093/nar/gkae402. PMID: 37961639 Free PMC article. Updated. Preprint.
References
-
- Kunkel T.A., Erie D.A.. DNA mismatch repair. Annu. Rev. Biochem. 2005; 74:681–710. - PubMed
-
- Au K.G., Welsh K., Modrich P.. Initiation of methyl-directed mismatch repair. J. Biol. Chem. 1992; 267:12142–12148. - PubMed
-
- Lahue R.S., Au K.G., Modrich P.. DNA mismatch correction in a defined system. Science. 1989; 245:160–164. - PubMed
-
- Cooper D.L., Lahue R.S., Modrich P.. Methyl-directed mismatch repair is bidirectional. J. Biol. Chem. 1993; 268:11823–11829. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

