ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis
- PMID: 28053229
- PMCID: PMC5255614
- DOI: 10.1073/pnas.1616299114
ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis
Abstract
Autophagy is a conserved pathway for bulk degradation of cytoplasmic material by a double-membrane structure named the autophagosome. The initiation of autophagosome formation requires the recruitment of autophagy-related protein 9 (ATG9) vesicles to the preautophagosomal structure. However, the functional relationship between ATG9 vesicles and the phagophore is controversial in different systems, and the molecular function of ATG9 remains unknown in plants. Here, we demonstrate that ATG9 is essential for endoplasmic reticulum (ER)-derived autophagosome formation in plants. Through a combination of genetic, in vivo imaging and electron tomography approaches, we show that Arabidopsis ATG9 deficiency leads to a drastic accumulation of autophagosome-related tubular structures in direct membrane continuity with the ER upon autophagic induction. Dynamic analyses demonstrate a transient membrane association between ATG9 vesicles and the autophagosomal membrane during autophagy. Furthermore, trafficking of ATG18a is compromised in atg9 mutants during autophagy by forming extended tubules in a phosphatidylinositol 3-phosphate-dependent manner. Taken together, this study provides evidence for a pivotal role of ATG9 in regulating autophagosome progression from the ER membrane in Arabidopsis.
Keywords: ATG18; ATG9; autophagosome; autophagy; endoplasmic reticulum.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
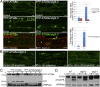


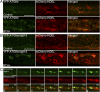

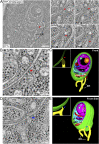
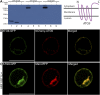


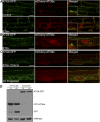
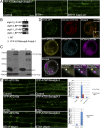

References
-
- Lamb CA, Yoshimori T, Tooze SA. The autophagosome: Origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14(12):759–774. - PubMed
-
- Michaeli S, Galili G, Genschik P, Fernie AR, Avin-Wittenberg T. Autophagy in plants: What’s new on the menu? Trends Plant Sci. 2016;21(2):134–144. - PubMed
-
- Liu Y, Bassham DC. Autophagy: Pathways for self-eating in plant cells. Annu Rev Plant Biol. 2012;63:215–237. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

