Innate and adaptive immune traits are differentially affected by genetic and environmental factors
- PMID: 28054551
- PMCID: PMC5227062
- DOI: 10.1038/ncomms13850
Innate and adaptive immune traits are differentially affected by genetic and environmental factors
Abstract
The diversity and activity of leukocytes is controlled by genetic and environmental influences to maintain balanced immune responses. However, the relative contribution of environmental compared with genetic factors that affect variations in immune traits is unknown. Here we analyse 23,394 immune phenotypes in 497 adult female twins. 76% of these traits show a predominantly heritable influence, whereas 24% are mostly influenced by environment. These data highlight the importance of shared childhood environmental influences such as diet, infections or microbes in shaping immune homeostasis for monocytes, B1 cells, γδ T cells and NKT cells, whereas dendritic cells, B2 cells, CD4+ T and CD8+ T cells are more influenced by genetics. Although leukocyte subsets are influenced by genetics and environment, adaptive immune traits are more affected by genetics, whereas innate immune traits are more affected by environment.
Figures

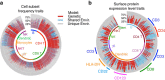
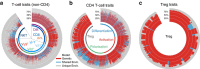


References
-
- Barreiro L. B. & Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat. Rev. Genet. 11, 17–30 (2010). - PubMed
-
- Ahmadi K. R. et al. Genetic determinism in the relationship between human CD4+ and CD8+ T lymphocyte populations? Genes Immun. 2, 381–387 (2001). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

