Attacked from All Sides: RNA Decay in Antiviral Defense
- PMID: 28054965
- PMCID: PMC5294971
- DOI: 10.3390/v9010002
Attacked from All Sides: RNA Decay in Antiviral Defense
Abstract
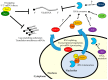
The innate immune system has evolved a number of sensors that recognize viral RNA (vRNA) to restrict infection, yet the full spectrum of host-encoded RNA binding proteins that target these foreign RNAs is still unknown. The RNA decay machinery, which uses exonucleases to degrade aberrant RNAs largely from the 5' or 3' end, is increasingly recognized as playing an important role in antiviral defense. The 5' degradation pathway can directly target viral messenger RNA (mRNA) for degradation, as well as indirectly attenuate replication by limiting specific pools of endogenous RNAs. The 3' degradation machinery (RNA exosome) is emerging as a downstream effector of a diverse array of vRNA sensors. This review discusses our current understanding of the roles of the RNA decay machinery in controlling viral infection.
Keywords: RNA decay; RNA-protein interactions; RNAse; TRAMP; Xrn1; antiviral; decapping; exonuclease; exosome; intrinsic immunity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Iwakawa H.-O., Tajima Y., Taniguchi T., Kaido M., Mise K., Tomari Y., Taniguchi H., Okuno T. Poly(A)-binding protein facilitates translation of an uncapped/nonpolyadenylated viral RNA by binding to the 3′ untranslated region. J. Virol. 2012;86:7836–7849. doi: 10.1128/JVI.00538-12. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

