Structural prediction of the interaction of the tumor suppressor p27KIP1 with cyclin A/CDK2 identifies a novel catalytically relevant determinant
- PMID: 28056778
- PMCID: PMC5217639
- DOI: 10.1186/s12859-016-1411-0
Structural prediction of the interaction of the tumor suppressor p27KIP1 with cyclin A/CDK2 identifies a novel catalytically relevant determinant
Abstract
Background: The cyclin-dependent kinase 2 (CDK2) together with its cyclin E and A partners is a central regulator of cell growth and division. Deregulation of CDK2 activity is associated with diseases such as cancer. The analysis of substrates identified S/T-P-X-R/K/H as the CDK2 consensus sequence. The crystal structure of cyclin A/CDK2 with a short model peptide supports this sequence and identifies key interactions. However, CDKs use additional determinants to recognize substrates, including the RXL motif that is read by the cyclin subunits. We were interested to determine whether additional amino acids beyond the minimal consensus sequence of the well-studied substrate and tumor suppressor p27KIP1 were relevant for catalysis.
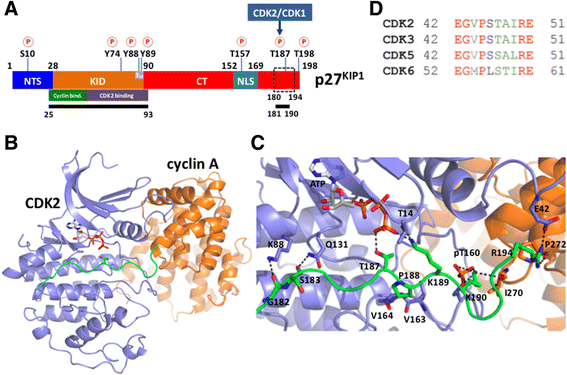
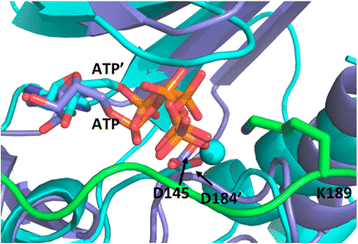
Results: To address whether additional amino acids, close to the minimal consensus sequence, play a role in binding, we investigate the interaction of cyclin A/CDK2 with an in vivo cellular partner and CDK inhibitor p27KIP1. This protein is an intrinsically unfolded protein and, in particular, the C-terminal half of the protein has not been accessible to structural analysis. This part harbors the CDK2 phosphorylation site. We used bioinformatics tools, including MODELLER, iTASSER and HADDOCK, along with partial structural information to build a model of the C-terminal region of p27KIP1 with cyclin A/CDK2. This revealed novel interactions beyond the consensus sequence with a proline and a basic amino acid at the P + 1 and the P + 3 sites, respectively. We suggest that the lysine at P + 2 might regulate the reversible association of the second counter ion in the active site of CDK2. The arginine at P + 7 interacts with both cyclin A and CDK2 and is important for the catalytic turnover rate.
Conclusion: Our modeling identifies additional amino acids in p27KIP1 beyond the consensus sequence that contribute to the efficiency of substrate phosphorylation.
Keywords: Cyclin-dependent kinase; Molecular modeling; Phosphorylation; Tumor suppressor; p27KIP1.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

