KinMap: a web-based tool for interactive navigation through human kinome data
- PMID: 28056780
- PMCID: PMC5217312
- DOI: 10.1186/s12859-016-1433-7
KinMap: a web-based tool for interactive navigation through human kinome data
Abstract
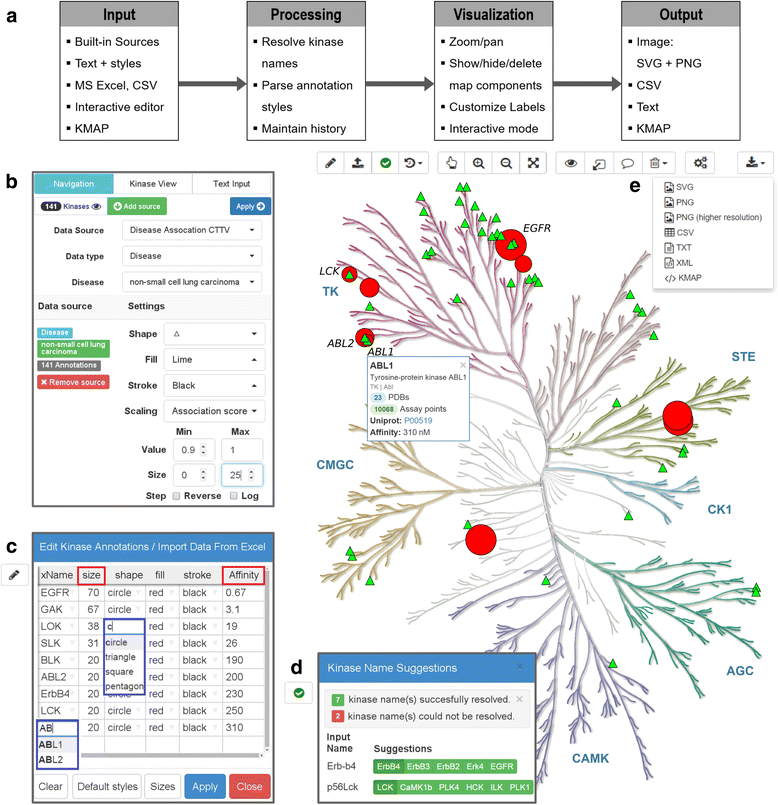
Background: Annotations of the phylogenetic tree of the human kinome is an intuitive way to visualize compound profiling data, structural features of kinases or functional relationships within this important class of proteins. The increasing volume and complexity of kinase-related data underlines the need for a tool that enables complex queries pertaining to kinase disease involvement and potential therapeutic uses of kinase inhibitors.
Results: Here, we present KinMap, a user-friendly online tool that facilitates the interactive navigation through kinase knowledge by linking biochemical, structural, and disease association data to the human kinome tree. To this end, preprocessed data from freely-available sources, such as ChEMBL, the Protein Data Bank, and the Center for Therapeutic Target Validation platform are integrated into KinMap and can easily be complemented by proprietary data. The value of KinMap will be exemplarily demonstrated for uncovering new therapeutic indications of known kinase inhibitors and for prioritizing kinases for drug development efforts.
Conclusion: KinMap represents a new generation of kinome tree viewers which facilitates interactive exploration of the human kinome. KinMap enables generation of high-quality annotated images of the human kinome tree as well as exchange of kinome-related data in scientific communications. Furthermore, KinMap supports multiple input and output formats and recognizes alternative kinase names and links them to a unified naming scheme, which makes it a useful tool across different disciplines and applications. A web-service of KinMap is freely available at http://www.kinhub.org/kinmap/ .
Keywords: Human kinome tree; Images; Interactive annotation; Protein kinases.
Figures

References
-
- Klebl B, Müller G, Hamacher M. Protein kinases as drug targets. Weinheim: Wiley-VCH; 2011. p. 49.
-
- Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, Floyd M, Hunt JP, Lockhart DJ, Milanov ZV, Morrison MJ, Pallares G, Patel HK, Pritchard S, Wodicka LM, Zarrinkar PP. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26(1):127–32. doi: 10.1038/nbt1358. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

