Satellite DNA and Transposable Elements in Seabuckthorn (Hippophae rhamnoides), a Dioecious Plant with Small Y and Large X Chromosomes
- PMID: 28057732
- PMCID: PMC5381607
- DOI: 10.1093/gbe/evw303
Satellite DNA and Transposable Elements in Seabuckthorn (Hippophae rhamnoides), a Dioecious Plant with Small Y and Large X Chromosomes
Abstract
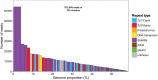
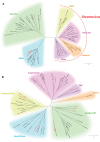
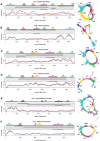
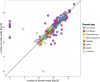
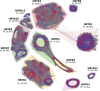
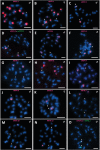
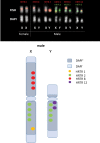
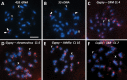
Seabuckthorn (Hippophae rhamnoides) is a dioecious shrub commonly used in the pharmaceutical, cosmetic, and environmental industry as a source of oil, minerals and vitamins. In this study, we analyzed the transposable elements and satellites in its genome. We carried out Illumina DNA sequencing and reconstructed the main repetitive DNA sequences. For data analysis, we developed a new bioinformatics approach for advanced satellite DNA analysis and showed that about 25% of the genome consists of satellite DNA and about 24% is formed of transposable elements, dominated by Ty3/Gypsy and Ty1/Copia LTR retrotransposons. FISH mapping revealed X chromosome-accumulated, Y chromosome-specific or both sex chromosomes-accumulated satellites but most satellites were found on autosomes. Transposable elements were located mostly in the subtelomeres of all chromosomes. The 5S rDNA and 45S rDNA were localized on one autosomal locus each. Although we demonstrated the small size of the Y chromosome of the seabuckthorn and accumulated satellite DNA there, we were unable to estimate the age and extent of the Y chromosome degeneration. Analysis of dioecious relatives such as Shepherdia would shed more light on the evolution of these sex chromosomes.
Keywords: chromosomal localization; genome composition; repetitive DNA; sex chromosomes.
© The Author(s) 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
References
-
- Alexandrov OS, Karlov GI. 2016. Molecular cytogenetic analysis and genomic organization of major DNA repeats in castor bean (Ricinus communis L.). Mol Genet Genomics 291:775–787. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

