PRMT2 interacts with splicing factors and regulates the alternative splicing of BCL-X
- PMID: 28057797
- PMCID: PMC5892402
- DOI: 10.1093/jb/mvw102
PRMT2 interacts with splicing factors and regulates the alternative splicing of BCL-X
Abstract
Protein arginine N-methyltransferase 2 (PRMT2) functions in JAK-STAT and Wnt/β-catenin signalling pathways, serves as a nuclear receptor-dependent transcriptional co-activator, and represses NF-κB and E2F1 transcription factor activities to promote apoptosis. We have previously demonstrated that PRMT2 interacts with PRMT1 and increases its activity. Here, we reveal associations using proteomics between the PRMT2 SH3 domain and splicing factors including Src-associated in mitosis 68 kDa protein (SAM68), a PRMT1 substrate and trans-acting factor that mediates BCL-X alternative splicing. We determined that PRMT2 interacts with SAM68 in cells and regulates its subcellular localization via the SH3 domain of PRMT2, prompting us to investigate the potential role of PRMT2 in BCL-X alternative splicing. We found that the expression of the full-length, wildtype form of PRMT2 promotes an increase in the BCL-X(L)/BCL-X(s) ratio in TNF-α or LPS stimulated cells. These results indicate that active PRMT2 may play a role during inflammation in alternative splicing regulation.
Keywords: BCL-X; PRMT2; SAM68; SH3 domain; alternative splicing.
© The Authors 2017. Published by Oxford University Press on behalf of the Japanese Biochemical Society. All rights reserved.
Figures
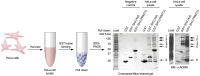
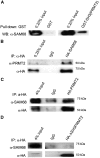
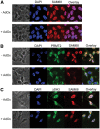
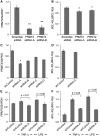
References
-
- Lin W.J., Gary J.D., Yang M.C., Clarke S., Herschman H.R. (1996) The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem. 271, 15034–15044 - PubMed
-
- Lakowski T.M., Frankel A. (2009) Kinetic analysis of human protein arginine N-methyltransferase 2: formation of monomethyl- and asymmetric dimethyl-arginine residues on histone H4. Biochem. J. 421, 253–261 - PubMed
-
- Tang J., Gary J.D., Clarke S., Herschman H.R. (1998) PRMT 3, a type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity, and regulation. J. Biol. Chem. 273, 16935–16945 - PubMed
-
- Chen D., Ma H., Hong H., Koh S.S., Huang S.M., Schurter B.T., Aswad D.W., Stallcup M.R. (1999) Regulation of transcription by a protein methyltransferase. Science 284, 2174–2177. - PubMed
-
- Schurter B.T., Koh S.S., Chen D., Bunick G.J., Harp J.M., Hanson B.L., Henschen-Edman A., Mackay D.R., Stallcup M.R., Aswad D.W. (2001) Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry 40, 5747–5756. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

