Aging related methylation influences the gene expression of key control genes in colorectal cancer and adenoma
- PMID: 28058013
- PMCID: PMC5175245
- DOI: 10.3748/wjg.v22.i47.10325
Aging related methylation influences the gene expression of key control genes in colorectal cancer and adenoma
Abstract
Aim: To analyze colorectal carcinogenesis and age-related DNA methylation alterations of gene sequences associated with epigenetic clock CpG sites.
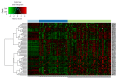
Methods: In silico DNA methylation analysis of 353 epigenetic clock CpG sites published by Steve Horvath was performed using methylation array data for a set of 123 colonic tissue samples [64 colorectal cancer (CRC), 42 adenoma, 17 normal; GEO accession number: GSE48684]. Among the differentially methylated age-related genes, secreted frizzled related protein 1 (SFRP1) promoter methylation was further investigated in colonic tissue from 8 healthy adults, 19 normal children, 20 adenoma and 8 CRC patients using bisulfite-specific PCR followed by methylation-specific high resolution melting (MS-HRM) analysis. mRNA expression of age-related "epigenetic clock" genes was studied using Affymetrix HGU133 Plus2.0 whole transcriptome data of 153 colonic biopsy samples (49 healthy adult, 49 adenoma, 49 CRC, 6 healthy children) (GEO accession numbers: GSE37364, GSE10714, GSE4183, GSE37267). Whole promoter methylation analysis of genes showing inverse DNA methylation-gene expression data was performed on 30 colonic samples using methyl capture sequencing.
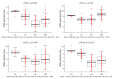
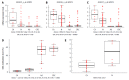
Results: Fifty-seven age-related CpG sites including hypermethylated PPP1R16B, SFRP1, SYNE1 and hypomethylated MGP, PIPOX were differentially methylated between CRC and normal tissues (P < 0.05, Δβ ≥ 10%). In the adenoma vs normal comparison, 70 CpG sites differed significantly, including hypermethylated DKK3, SDC2, SFRP1, SYNE1 and hypomethylated CEMIP, SPATA18 (P < 0.05, Δβ ≥ 10%). In MS-HRM analysis, the SFRP1 promoter region was significantly hypermethylated in CRC (55.0% ± 8.4 %) and adenoma tissue samples (49.9% ± 18.1%) compared to normal adult (5.2% ± 2.7%) and young (2.2% ± 0.7%) colonic tissue (P < 0.0001). DNA methylation of SFRP1 promoter was slightly, but significantly increased in healthy adults compared to normal young samples (P < 0.02). This correlated with significantly increased SFRP1 mRNA levels in children compared to normal adult samples (P < 0.05). In CRC tissue the mRNA expression of 117 age-related genes were changed, while in adenoma samples 102 genes showed differential expression compared with normal colonic tissue (P < 0.05, logFC > 0.5). The change of expression for several genes including SYNE1, CLEC3B, LTBP3 and SFRP1, followed the same pattern in aging and carcinogenesis, though not for all genes (e.g., MGP).
Conclusion: Several age-related DNA methylation alterations can be observed during CRC development and progression affecting the mRNA expression of certain CRC- and adenoma-related key control genes.
Keywords: Adenoma; Aging; Colorectal cancer; DNA methylation; Epigenetic clock; Epigenetic drift; Secreted frizzled related protein 1.
Conflict of interest statement
Conflict-of-interest statement: The authors declare that no conflict of interest exists.
Figures

References
-
- Finkel T, Serrano M, Blasco MA. The common biology of cancer and ageing. Nature. 2007;448:767–774. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

