SHG-specificity of cellular Rootletin filaments enables naïve imaging with universal conservation
- PMID: 28059168
- PMCID: PMC5216331
- DOI: 10.1038/srep39967
SHG-specificity of cellular Rootletin filaments enables naïve imaging with universal conservation
Abstract
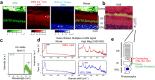
Despite growing demand for truly naïve imaging, label-free observation of cilium-related structure remains challenging, and validation of the pertinent molecules is correspondingly difficult. In this study, in retinas and cultured cells, we distinctively visualized Rootletin filaments in rootlets in the second harmonic generation (SHG) channel, integrated in custom coherent nonlinear optical microscopy (CNOM) with a simple, compact, and ultra-broadband supercontinuum light source. This SHG signal was primarily detected on rootlets of connecting cilia in the retinal photoreceptor and was validated by colocalization with anti-Rootletin staining. Transfection of cells with Rootletin fragments revealed that the SHG signal can be ascribed to filaments assembled from the R234 domain, but not to cross-striations assembled from the R123 domain. Consistent with this, Rootletin-depleted cells lacked SHG signal expected as centrosome linker. As a proof of concept, we confirmed that similar fibrous SHG was observed even in unicellular ciliates. These findings have potential for broad applications in clinical diagnosis and biophysical experiments with various organisms.
Figures




References
-
- Muller M. & Schins J. M. Imaging the thermodynamic state of lipid membranes with multiplex CARS microscopy. J. Phys. Chem. B 106, 3715–3723 (2002).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

