Structural and functional alterations in the colonic microbiome of the rat in a model of stress induced irritable bowel syndrome
- PMID: 28059627
- PMCID: PMC5341915
- DOI: 10.1080/19490976.2016.1273999
Structural and functional alterations in the colonic microbiome of the rat in a model of stress induced irritable bowel syndrome
Abstract
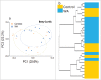
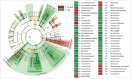
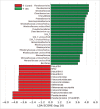
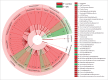
Stress is known to perturb the microbiome and exacerbate irritable bowel syndrome (IBS) associated symptoms. Characterizing structural and functional changes in the microbiome is necessary to understand how alterations affect the biomolecular environment of the gut in IBS. Repeated water avoidance (WA) stress was used to induce IBS-like symptoms in rats. The colon-mucosa associated microbiome was characterized in 13 stressed and control animals by 16S sequencing. In silico analysis of the functional domains of microbial communities was done by inferring metagenomic profiles from 16S data. Microbial communities and functional profiles were compared between conditions. WA animals exhibited higher α-diversity and moderate divergence in community structure (β-diversity) compared with controls. Specific clades and taxa were consistently and significantly modified in the WA animals. The WA microbiome was particularly enriched in Proteobacteria and depleted in several beneficial taxa. A decreased capacity in metabolic domains, including energy- and lipid-metabolism, and an increased capacity for fatty acid and sulfur metabolism was inferred for the WA microbiome. The stressed condition favored the proliferation of a greater diversity of microbes that appear to be functionally similar, resulting in a functionally poorer microbiome with implications for epithelial health. Taxa, with known beneficial effects, were found to be depleted, which supports their relevance as therapeutic agents to restore microbial health. Microbial sulfur metabolism may form a key component of visceral nerve sensitization pathways and is therefore of interest as a target metabolic domain in microbial ecological restoration.
Keywords: colon; microbiome; mucosa; stress; visceral hypersensitivity.
Figures



Similar articles
-
The microbiome of the oral mucosa in irritable bowel syndrome.Gut Microbes. 2016 Jul 3;7(4):286-301. doi: 10.1080/19490976.2016.1162363. Epub 2016 Mar 10. Gut Microbes. 2016. PMID: 26963804 Free PMC article.
-
Differences in gut microbial composition correlate with regional brain volumes in irritable bowel syndrome.Microbiome. 2017 May 1;5(1):49. doi: 10.1186/s40168-017-0260-z. Microbiome. 2017. PMID: 28457228 Free PMC article.
-
Effect of mild moxibustion on intestinal microbiota and NLRP6 inflammasome signaling in rats with post-inflammatory irritable bowel syndrome.World J Gastroenterol. 2019 Aug 28;25(32):4696-4714. doi: 10.3748/wjg.v25.i32.4696. World J Gastroenterol. 2019. PMID: 31528095 Free PMC article.
-
Overgrowth of the indigenous gut microbiome and irritable bowel syndrome.World J Gastroenterol. 2014 Mar 14;20(10):2449-55. doi: 10.3748/wjg.v20.i10.2449. World J Gastroenterol. 2014. PMID: 24627582 Free PMC article. Review.
-
A Microbial Relationship Between Irritable Bowel Syndrome and Depressive Symptoms.Biol Res Nurs. 2021 Jan;23(1):50-64. doi: 10.1177/1099800420940787. Epub 2020 Jul 24. Biol Res Nurs. 2021. PMID: 32705884 Review.
Cited by
-
Animal models of visceral pain and the role of the microbiome.Neurobiol Pain. 2021 May 28;10:100064. doi: 10.1016/j.ynpai.2021.100064. eCollection 2021 Aug-Dec. Neurobiol Pain. 2021. PMID: 34151049 Free PMC article. Review.
-
Transcriptome and methylome profiling in a rat model of irritable bowel syndrome induced by stress.Int J Mol Med. 2018 Nov;42(5):2641-2649. doi: 10.3892/ijmm.2018.3823. Epub 2018 Aug 14. Int J Mol Med. 2018. PMID: 30106160 Free PMC article.
-
Microbial imbalance in Chinese children with diarrhea or constipation.Sci Rep. 2024 Jun 12;14(1):13516. doi: 10.1038/s41598-024-60683-6. Sci Rep. 2024. PMID: 38866797 Free PMC article.
-
Metagenomics and metabolomics analysis to investigate the effect of Shugan decoction on intestinal microbiota in irritable bowel syndrome rats.Front Microbiol. 2022 Nov 21;13:1024822. doi: 10.3389/fmicb.2022.1024822. eCollection 2022. Front Microbiol. 2022. PMID: 36478867 Free PMC article.
-
The ethanol extract of Periplaneta Americana L. improves ulcerative colitis induced by a combination of chronic stress and TNBS in rats.Acta Cir Bras. 2022 Aug 12;37(5):e370505. doi: 10.1590/acb370505. eCollection 2022. Acta Cir Bras. 2022. PMID: 35976342 Free PMC article.
References
-
- Mayer EA, Collins SM. Evolving pathophysiologic models of functional gastrointestinal disorders. Gastroenterology 2002; 122:2032-48; PMID:12055608; http://dx.doi.org/10.1053/gast.2002.33584 - DOI - PubMed
-
- Kalmokoff M, Franklin J, Petronella N, Green J, Brooks SP. Phylum level change in the cecal and fecal gut communities of rats fed diets containing different fermentable substrates supports a role for nitrogen as a factor contributing to community structure. Nutrients 2015; 7:3279-99; PMID:25954902; http://dx.doi.org/10.3390/nu7053279 - DOI - PMC - PubMed
-
- Wos-Oxley ML, Bleich A, Oxley AP, Kahl S, Janus LM, Smoczek A, Nahrstedt H, Pils MC, Taudien S, Platzer M, et al.. Comparative evaluation of establishing a human gut microbial community within rodent models. Gut Microbes 2012; 3:234-49; PMID:22572831; http://dx.doi.org/10.4161/gmic.19934 - DOI - PMC - PubMed
-
- Jiminez JA, Uwiera TC, Inglis GD, Uwiera RR. Animal models to study acute and chronic intestinal inflammation in mammals. Gut Pathogens 2015; 7:1; PMID:25653718; http://dx.doi.org/10.1186/s13099-015-0076-y - DOI - PMC - PubMed
-
- Hong S, Fan J, Kemmerer ES, Evans S, Li Y, Wiley JW. Reciprocal changes in vanilloid (TRPV1) and endocannabinoid (CB1) receptors contribute to visceral hyperalgesia in the water avoidance stressed Rat. Gut 2009; 58:202-10; PMID:18936104; http://dx.doi.org/10.1136/gut.2008.157594 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
