Past1 Modulates Drosophila Eye Development
- PMID: 28060904
- PMCID: PMC5218476
- DOI: 10.1371/journal.pone.0169639
Past1 Modulates Drosophila Eye Development
Erratum in
-
Correction: Past1 Modulates Drosophila Eye Development.PLoS One. 2017 Mar 20;12(3):e0174495. doi: 10.1371/journal.pone.0174495. eCollection 2017. PLoS One. 2017. PMID: 28319181 Free PMC article.
Abstract
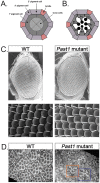
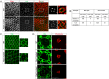
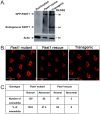
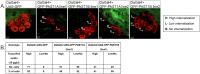
Endocytosis is a multi-step process involving a large number of proteins, both general factors, such as clathrin and adaptor protein complexes, and unique proteins, which modulate specialized endocytic processes, like the EHD proteins. EHDs are a family of Eps15 Homology Domain containing proteins that consists of four mammalian homologs, one C. elegans, one Drosophila melanogaster and two plants orthologs. These membrane-associated proteins are involved in different steps of endocytic trafficking pathways. We have previously shown that the Drosophila EHD ortholog, PAST1, associates predominantly with the plasma membrane. Mutations in Past1 result in defects in endocytosis, male sterility, temperature sensitivity and premature death of the flies. Also, Past1 genetically interacts with Notch. In the present study, we investigated the role of PAST1 in the developing fly eye. In mutant flies lacking PAST1, abnormal differentiation of photoreceptors R1, R6 and R7 was evident, with partial penetrance. Likewise, five cone cells were present instead of four. Expression of transgenic PAST1 resulted in a dominant negative effect, with a phenotype similar to that of the deletion mutant, and appearance of additional inter-ommatidial pigment cells. Our results strongly suggest a role for PAST1 in differentiation of photoreceptors R1/R6/R7 and cone cells of the fly ommatidia.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







Similar articles
-
Drosophila Past1 is involved in endocytosis and is required for germline development and survival of the adult fly.J Cell Sci. 2009 Feb 15;122(Pt 4):471-80. doi: 10.1242/jcs.038521. Epub 2009 Jan 27. J Cell Sci. 2009. PMID: 19174465
-
The EHD protein Past1 controls postsynaptic membrane elaboration and synaptic function.Mol Biol Cell. 2015 Sep 15;26(18):3275-88. doi: 10.1091/mbc.E15-02-0093. Epub 2015 Jul 22. Mol Biol Cell. 2015. PMID: 26202464 Free PMC article.
-
Shared as well as distinct roles of EHD proteins revealed by biochemical and functional comparisons in mammalian cells and C. elegans.BMC Cell Biol. 2007 Jan 18;8:3. doi: 10.1186/1471-2121-8-3. BMC Cell Biol. 2007. PMID: 17233914 Free PMC article.
-
Signaling mechanisms in induction of the R7 photoreceptor in the developing Drosophila retina.Bioessays. 1994 Apr;16(4):237-44. doi: 10.1002/bies.950160406. Bioessays. 1994. PMID: 8031300 Review.
-
The role of EHD proteins at the neuronal synapse.Sci Signal. 2012 Apr 24;5(221):jc1. doi: 10.1126/scisignal.2002989. Sci Signal. 2012. PMID: 22534130 Review.
Cited by
-
Regulators of cell movement during development and regeneration in Drosophila.Open Biol. 2019 May 31;9(5):180245. doi: 10.1098/rsob.180245. Open Biol. 2019. PMID: 31039676 Free PMC article.
-
Correction: Past1 Modulates Drosophila Eye Development.PLoS One. 2017 Mar 20;12(3):e0174495. doi: 10.1371/journal.pone.0174495. eCollection 2017. PLoS One. 2017. PMID: 28319181 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

