The occurrence of ESBL-producing Escherichia coli carrying aminoglycoside resistance genes in urinary tract infections in Saudi Arabia
- PMID: 28061852
- PMCID: PMC5219782
- DOI: 10.1186/s12941-016-0177-6
The occurrence of ESBL-producing Escherichia coli carrying aminoglycoside resistance genes in urinary tract infections in Saudi Arabia
Abstract
Background: The infection and prevalence of extended-spectrum β-lactamases (ESBLs) is a worldwide problem, and the presence of ESBLs varies between countries. In this study, we investigated the occurrence of plasmid-mediated ESBL/AmpC/carbapenemase/aminoglycoside resistance gene expression in Escherichia coli using phenotypic and genotypic techniques.
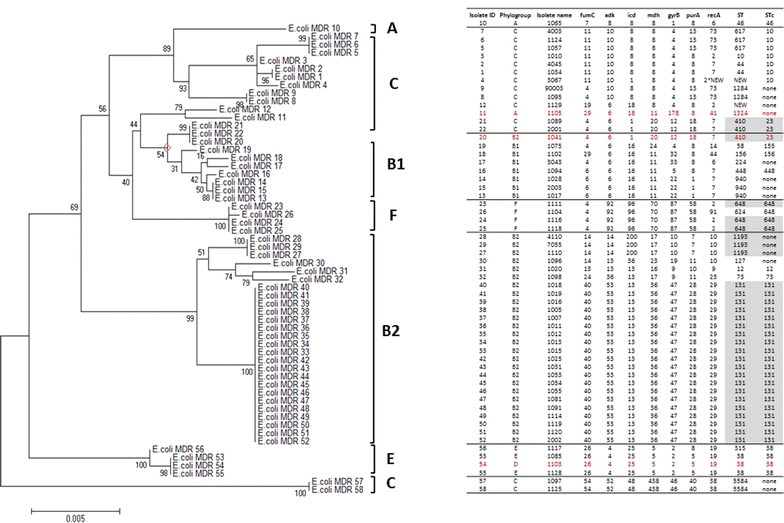
Methods: A total of 58 E. coli isolates were collected from hospitals in the city of Makkah and screened for the production of ESBL/AmpC/carbapenemase/aminoglycoside resistance genes. All samples were subjected to phenotypic and genotypic analyses. The antibiotic susceptibility of the E. coli isolates was determined using the Vitek-2 system and the minimum inhibitory concentration (MIC) assay. Antimicrobial agents tested using the Vitek 2 system and MIC assay included the expanded-spectrum (or third-generation) cephalosporins (e.g., cefoxitin, cefepime, aztreonam, cefotaxime, ceftriaxone, and ceftazidime) and carbapenems (meropenem and imipenem). Reported positive isolates were investigated using genotyping technology (oligonucleotide microarray-based assay and PCR). The genotyping investigation was focused on ESBL variants and the AmpC, carbapenemase and aminoglycoside resistance genes. E. coli was phylogenetically grouped, and the clonality of the isolates was studied using multilocus sequence typing (MLST).
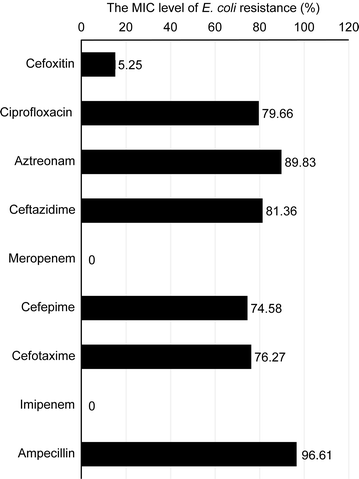
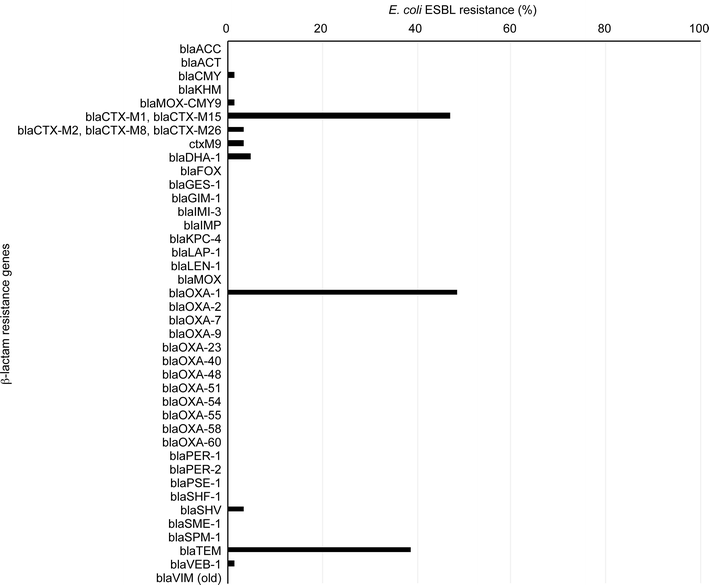
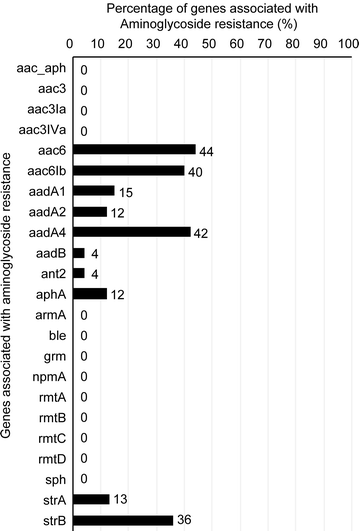
Results: Our E. coli isolates exhibited different levels of resistance to ESBL drugs, including ampicillin (96.61%), cefoxitin (15.25%), ciprofloxacin (79.66%), cefepime (75.58%), aztreonam (89.83%), cefotaxime (76.27%), ceftazidime (81.36%), meropenem (0%) and imipenem (0%). Furthermore, the distribution of ESBL-producing E. coli was consistent with the data obtained using an oligonucleotide microarray-based assay and PCR genotyping against genes associated with β-lactam resistance. ST131 was the dominant sequence type lineage of the isolates and was the most uropathogenic E. coli lineage. The E. coli isolates also carried aminoglycoside resistance genes.
Conclusions: The evolution and prevalence of ESBL-producing E. coli may be rapidly accelerating in Saudi Arabia due to the high visitation seasons (especially to the city of Makkah). The health authority in Saudi Arabia should monitor the level of drug resistance in all general hospitals to reduce the increasing trend of microbial drug resistance and the impact on patient therapy.
Keywords: E. coli; ESBL; Genotyping; K. pneumonia; Phenotyping; Saudi Arabia.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

